Difference between revisions of "GutR"
| Line 1: | Line 1: | ||
| − | + | ||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of glucitol utilization | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of glucitol utilization | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU06140 gutR] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism]''' | ||
Revision as of 15:35, 6 August 2012
| Gene name | gutR |
| Synonyms | |
| Essential | no |
| Product | transcription activator |
| Function | regulation of glucitol utilization |
| Gene expression levels in SubtiExpress: gutR | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 94 kDa, 6.113 |
| Gene length, protein length | 2487 bp, 829 aa |
| Immediate neighbours | ydjC, gutB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 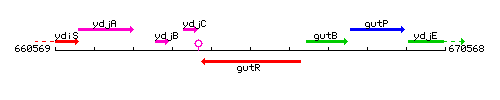
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control
This gene is a member of the following regulons
The GutR regulon:
The gene
Basic information
- Locus tag: BSU06140
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39143
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: gutR PubMed
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Shouji Watanabe, Miyuki Hamano, Hiroshi Kakeshita, Keigo Bunai, Shigeo Tojo, Hirotake Yamaguchi, Yasutaro Fujita, Sui-Lam Wong, Kunio Yamane
Mannitol-1-phosphate dehydrogenase (MtlD) is required for mannitol and glucitol assimilation in Bacillus subtilis: possible cooperation of mtl and gut operons.
J Bacteriol: 2003, 185(16);4816-24
[PubMed:12897001]
[WorldCat.org]
[DOI]
(P p)
K K Poon, J C Chu, S L Wong
Roles of glucitol in the GutR-mediated transcription activation process in Bacillus subtilis: glucitol induces GutR to change its conformation and to bind ATP.
J Biol Chem: 2001, 276(32);29819-25
[PubMed:11390381]
[WorldCat.org]
[DOI]
(P p)
K K Poon, C L Chen, S L Wong
Roles of glucitol in the GutR-mediated transcription activation process in Bacillus subtilis: tight binding of GutR to tis binding site.
J Biol Chem: 2001, 276(13);9620-5
[PubMed:11118449]
[WorldCat.org]
[DOI]
(P p)
R Ye, S N Rehemtulla, S L Wong
Glucitol induction in Bacillus subtilis is mediated by a regulatory factor, GutR.
J Bacteriol: 1994, 176(11);3321-7
[PubMed:8195087]
[WorldCat.org]
[DOI]
(P p)
R Ye, S L Wong
Transcriptional regulation of the Bacillus subtilis glucitol dehydrogenase gene.
J Bacteriol: 1994, 176(11);3314-20
[PubMed:8195086]
[WorldCat.org]
[DOI]
(P p)