Difference between revisions of "GpsB"
| Line 27: | Line 27: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=gpsB_2331779_2332075_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:gpsB_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=gpsB_2331779_2332075_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:gpsB_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU22180]] |
|- | |- | ||
|} | |} | ||
Revision as of 13:43, 16 May 2013
- Description: required for removal of PBP1 from the cell pole after completion of cell pole maturation
| Gene name | gpsB |
| Synonyms | ypsB |
| Essential | no |
| Product | cell division protein, member of the divisome |
| Function | cell division, cell elongation |
| Gene expression levels in SubtiExpress: gpsB | |
| MW, pI | 11 kDa, 5.629 |
| Gene length, protein length | 294 bp, 98 aa |
| Immediate neighbours | rnpB, ypsA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 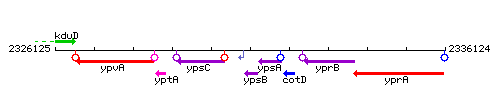
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell division, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22180
Phenotypes of a mutant
- synthetic lethal phenotype with a ftsA mutation PubMed; synthetic sick phenotype when combined with an ezrA mutation PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): DivIVA
Extended information on the protein
- Kinetic information:
- Domains: contains a DivIVA-like N-terminal domain
- Modification: phosphorylation on (Ser-73 OR Thr-75) PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P50839
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Pamela Gamba, Jan-Willem Veening, Nigel J Saunders, Leendert W Hamoen, Richard A Daniel
Two-step assembly dynamics of the Bacillus subtilis divisome.
J Bacteriol: 2009, 191(13);4186-94
[PubMed:19429628]
[WorldCat.org]
[DOI]
(I p)
José Roberto Tavares, Robson F de Souza, Guilherme Louzada Silva Meira, Frederico J Gueiros-Filho
Cytological characterization of YpsB, a novel component of the Bacillus subtilis divisome.
J Bacteriol: 2008, 190(21);7096-107
[PubMed:18776011]
[WorldCat.org]
[DOI]
(I p)
Dennis Claessen, Robyn Emmins, Leendert W Hamoen, Richard A Daniel, Jeff Errington, David H Edwards
Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis.
Mol Microbiol: 2008, 68(4);1029-46
[PubMed:18363795]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)