Difference between revisions of "GndA"
| Line 119: | Line 119: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 10759 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 15566 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:30, 17 April 2014
- Description: NADP-dependent phosphogluconate dehydrogenase
| Gene name | gndA |
| Synonyms | yqjI |
| Essential | no |
| Product | NADP-dependent phosphogluconate dehydrogenase |
| Function | pentose phosphate pathway |
| Gene expression levels in SubtiExpress: gndA | |
| Metabolic function and regulation of this protein in SubtiPathways: gndA | |
| MW, pI | 51 kDa, 5.076 |
| Gene length, protein length | 1407 bp, 469 aa |
| Immediate neighbours | zwf, polY1 |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 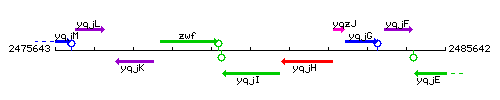
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23860
Phenotypes of a mutant
Database entries
- BsubCyc: BSU23860
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 6-phospho-D-gluconate + NADP+ = D-ribulose 5-phosphate + CO2 + NADPH (according to Swiss-Prot)
- Protein family: 6-phosphogluconate dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s): GntZ
Extended information on the protein
- Kinetic information: Reversible reaction PubMed
- Modification: phosphorylation on (Thr-15 OR Thr-17) PubMed
- Effectors of protein activity:
Database entries
- BsubCyc: BSU23860
- UniProt: P80859
- KEGG entry: [3]
- E.C. number: 1.1.1.44
Additional information
It contains a cysteine on the active site. The enzyme is a dimer. PubMed
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 10759 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 15566 PubMed
Biological materials
- Mutant: GP1514 (gndA::kan), available in Jörg Stülke's lab
- Expression vector:
- pGP1777 (for expression, purification in E. coli with N-terminal Strep-tag, in pGP172, available in Jörg Stülke's lab)
- pGP1789 (N-terminal Strep-tag, for SPINE, purification from B. subtilis, in pGP1389) (available in Jörg Stülke's lab)
- GP1408 (gndA-Strep (spc)) & GP1410 (gndA-Strep (cat)), purification from B. subtilis, for SPINE, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
- FLAG-tag construct:
- GP1403 (spc, based on pGP1331), available in Jörg Stülke's lab
- GP1408 (kan, resistance cassette exchange in GP1403), available in Jörg Stülke's lab
Labs working on this gene/protein
Your additional remarks
References
Scott Cameron, Viviane P Martini, Jorge Iulek, William N Hunter
Geobacillus stearothermophilus 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2009, 65(Pt 5);450-4
[PubMed:19407374]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Nicola Zamboni, Eliane Fischer, Dietmar Laudert, Stéphane Aymerich, Hans-Peter Hohmann, Uwe Sauer
The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway.
J Bacteriol: 2004, 186(14);4528-34
[PubMed:15231785]
[WorldCat.org]
[DOI]
(P p)