GmuG
- Description: beta-1,4-mannanase
| Gene name | gmuG |
| Synonyms | ydhT |
| Essential | no |
| Product | beta-1,4-mannanase |
| Function | glucomannan utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 40 kDa, 5.628 |
| Gene length, protein length | 1086 bp, 362 aa |
| Immediate neighbours | gmuF, ydhU |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 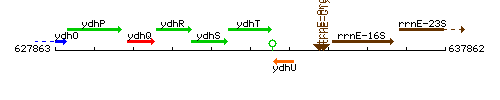
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
AbrB regulon, CcpA regulon, GmuR regulon
The gene
Basic information
- Locus tag: BSU05880
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Random hydrolysis of (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 26 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: secreted (according to Swiss-Prot), extracellular (signal peptide) PubMed
Database entries
- UniProt: O05512
- KEGG entry: [3]
- E.C. number: 3.2.1.78
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Christian Degering, Thorsten Eggert, Michael Puls, Johannes Bongaerts, Stefan Evers, Karl-Heinz Maurer, Karl-Erich Jaeger
Optimization of protease secretion in Bacillus subtilis and Bacillus licheniformis by screening of homologous and heterologous signal peptides.
Appl Environ Microbiol: 2010, 76(19);6370-6
[PubMed:20709850]
[WorldCat.org]
[DOI]
(I p)
Jiayun Qiao, Zhenghua Rao, Bing Dong, Yunhe Cao
Expression of Bacillus subtilis MA139 beta-mannanase in Pichia pastoris and the enzyme characterization.
Appl Biochem Biotechnol: 2010, 160(5);1362-70
[PubMed:19504356]
[WorldCat.org]
[DOI]
(I p)
Louise E Tailford, Valerie M-A Ducros, James E Flint, Shirley M Roberts, Carl Morland, David L Zechel, Nicola Smith, Mads E Bjørnvad, Torben V Borchert, Keith S Wilson, Gideon J Davies, Harry J Gilbert
Understanding how diverse beta-mannanases recognize heterogeneous substrates.
Biochemistry: 2009, 48(29);7009-18
[PubMed:19441796]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Yoshito Sadaie, Hisashi Nakadate, Reiko Fukui, Lii Mien Yee, Kei Asai
Glucomannan utilization operon of Bacillus subtilis.
FEMS Microbiol Lett: 2008, 279(1);103-9
[PubMed:18177310]
[WorldCat.org]
[DOI]
(P p)