Difference between revisions of "GlyS"
(→Expression and regulation) |
(→Database entries) |
||
| Line 82: | Line 82: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1J5W 1J5W] (from ''Thermotoga maritima'', 55% identity, 72% similarity) |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P54381 P54381] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P54381 P54381] | ||
Revision as of 17:51, 18 February 2010
- Description: glycyl-tRNA synthetase (beta subunit)
| Gene name | glyS |
| Synonyms | yqfK |
| Essential | yes PubMed |
| Product | glycyl-tRNA synthetase (beta subunit) |
| Function | translation |
| Metabolic function and regulation of this protein in SubtiPathways: tRNA charging | |
| MW, pI | 76 kDa, 4.973 |
| Gene length, protein length | 2037 bp, 679 aa |
| Immediate neighbours | ccpN, glyQ |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 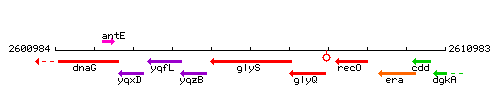
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU25260
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly) (according to Swiss-Prot)
- Protein family: class-II aminoacyl-tRNA synthetase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 1J5W (from Thermotoga maritima, 55% identity, 72% similarity)
- UniProt: P54381
- KEGG entry: [3]
- E.C. number: 6.1.1.14
Additional information
Expression and regulation
- Regulatory mechanism:
- T-box: RNA switch, transcriptional antitermination PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
Audrey R Nelson, Tina M Henkin, Paul F Agris
tRNA regulation of gene expression: interactions of an mRNA 5'-UTR with a regulatory tRNA.
RNA: 2006, 12(7);1254-61
[PubMed:16741230]
[WorldCat.org]
[DOI]
(P p)
Mary R Yousef, Frank J Grundy, Tina M Henkin
Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA.
J Mol Biol: 2005, 349(2);273-87
[PubMed:15890195]
[WorldCat.org]
[DOI]
(P p)
Frank J Grundy, Mary R Yousef, Tina M Henkin
Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene.
J Mol Biol: 2005, 346(1);73-81
[PubMed:15663928]
[WorldCat.org]
[DOI]
(P p)
Frank J Grundy, Tina M Henkin
Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro.
J Bacteriol: 2004, 186(16);5392-9
[PubMed:15292140]
[WorldCat.org]
[DOI]
(P p)