Difference between revisions of "GlyA"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[biosynthesis/ acquisition of amino acids]]}}, | {{SubtiWiki category|[[biosynthesis/ acquisition of amino acids]]}}, | ||
{{SubtiWiki category|[[essential genes]]}}, | {{SubtiWiki category|[[essential genes]]}}, | ||
| − | {{SubtiWiki category|[[phosphoproteins]]}} | + | {{SubtiWiki category|[[phosphoproteins]]}}, |
| + | [[most abundant proteins]] | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 69: | Line 66: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 87: | Line 81: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylated on ser/ thr/ tyr [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed] | * '''Modification:''' phosphorylated on ser/ thr/ tyr [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed] | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 117: | Line 111: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=glyA_3789190_3790437_-1 glyA] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=glyA_3789190_3790437_-1 glyA] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|11591660}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|11591660}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 128: | Line 122: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
=Biological materials = | =Biological materials = | ||
| Line 149: | Line 144: | ||
=References= | =References= | ||
| − | <pubmed>19258532,11591660, 20152942,11591660, 19171795, 17726680, 16493705 </pubmed> | + | <pubmed>19258532,11591660, 20152942,11591660, 19171795, 17726680, 16493705 15378759</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:00, 5 March 2014
- Description: serine hydroxymethyltransferase
| Gene name | glyA |
| Synonyms | glyC, ipc-34d |
| Essential | yes PubMed |
| Product | serine hydroxymethyltransferase |
| Function | biosynthesis of glycine |
| Gene expression levels in SubtiExpress: glyA | |
| Metabolic function and regulation of this protein in SubtiPathways: glyA | |
| MW, pI | 45 kDa, 5.475 |
| Gene length, protein length | 1245 bp, 415 aa |
| Immediate neighbours | upp, ywlG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 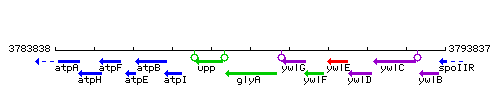
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, essential genes, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU36900
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 5,10-methylenetetrahydrofolate + glycine + H2O = tetrahydrofolate + L-serine (according to Swiss-Prot)
- Protein family: SHMT family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 2VGU (complex with L-serine, Geobacillus stearothermophilus), 2VI8 (Geobacillus stearothermophilus)
- UniProt: P39148
- KEGG entry: [3]
- E.C. number: 2.1.2.1
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- T-box: RNA switch, transcriptional antitermination PubMed
- PurR: transcription repression (molecular inducer: PRPP) PubMed
- Additional information:
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Anant Narayan Bhatt, Vinod Bhakuni, Ashutosh Kumar, M Yahiya Khan, Mohammad Imran Siddiqi
Alkaline pH-dependent differential unfolding characteristics of mesophilic and thermophilic homologs of dimeric serine hydroxymethyltransferase.
Biochim Biophys Acta: 2010, 1804(6);1294-300
[PubMed:20152942]
[WorldCat.org]
[DOI]
(P p)
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
Yann Duroc, Carmela Giglione, Thierry Meinnel
Mutations in three distinct loci cause resistance to peptide deformylase inhibitors in Bacillus subtilis.
Antimicrob Agents Chemother: 2009, 53(4);1673-8
[PubMed:19171795]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
H H Saxild, K Brunstedt, K I Nielsen, H Jarmer, P Nygaard
Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO.
J Bacteriol: 2001, 183(21);6175-83
[PubMed:11591660]
[WorldCat.org]
[DOI]
(P p)