Difference between revisions of "GltT"
| Line 36: | Line 36: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 98: | Line 94: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.pdb.org/pdb/explore/remediatedSequence.do?structureId=4IZM 4IZM] (the protein from ''Pyrococcus horikoshii'', 35% identity, 73% similarity) {{PubMed|23563139}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O07605 O07605] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O07605 O07605] | ||
| Line 142: | Line 138: | ||
=References= | =References= | ||
| − | <pubmed>18763711, </pubmed> | + | <pubmed>18763711, 23563139 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:58, 20 August 2013
- Description: major H+/Na+-glutamate symport protein
| Gene name | gltT |
| Synonyms | yhfG |
| Essential | no |
| Product | major H+/Na+-glutamate symport protein |
| Function | glutamate and aspartate uptake |
| Gene expression levels in SubtiExpress: gltT | |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 45 kDa, 9.029 |
| Gene length, protein length | 1287 bp, 429 aa |
| Immediate neighbours | yhfF, yhfH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 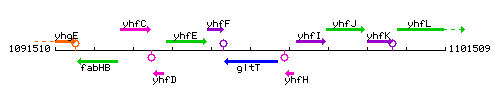
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed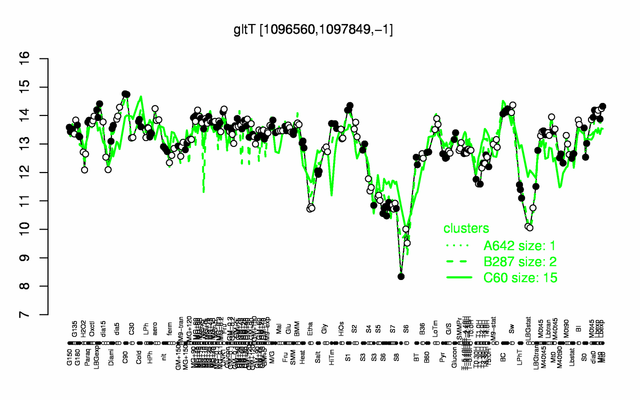
| |
Contents
Categories containing this gene/protein
transporters/ other, biosynthesis/ acquisition of amino acids, glutamate metabolism, utilization of amino acids, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU10220
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: View classification (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane PubMed
Database entries
- UniProt: O07605
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Nicolas Reyes, SeCheol Oh, Olga Boudker
Binding thermodynamics of a glutamate transporter homolog.
Nat Struct Mol Biol: 2013, 20(5);634-40
[PubMed:23563139]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)