Difference between revisions of "GlmM"
| Line 122: | Line 122: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** A [[ncRNA]] is predicted between ''[[glmM]]'' and ''[[glmS]]'' {{PubMed|20525796}} | ** A [[ncRNA]] is predicted between ''[[glmM]]'' and ''[[glmS]]'' {{PubMed|20525796}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 1353 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 2093 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:30, 17 April 2014
- Description: phosphoglucosamine mutase, required for cell wall synthesis
| Gene name | glmM |
| Synonyms | ybbT |
| Essential | yes PubMed |
| Product | phosphoglucosamine mutase, required for cell wall synthesis |
| Function | cell wall synthesis |
| Gene expression levels in SubtiExpress: glmM | |
| Metabolic function and regulation of this protein in SubtiPathways: glmM | |
| MW, pI | 48 kDa, 4.677 |
| Gene length, protein length | 1344 bp, 448 aa |
| Immediate neighbours | cdaR, glmS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 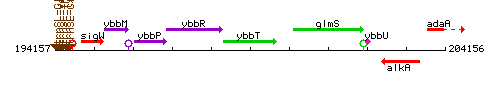
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed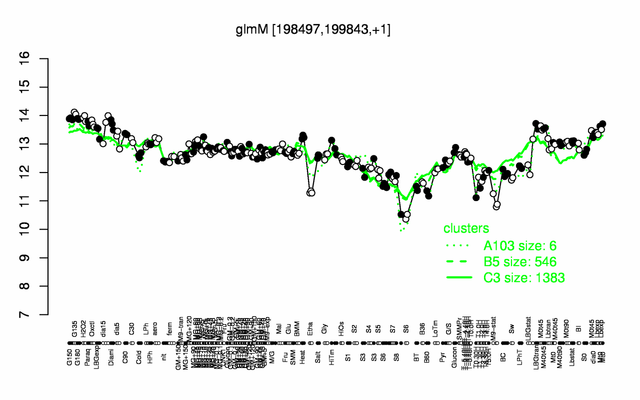
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, essential genes, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01770
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU01770
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Alpha-D-glucosamine 1-phosphate = D-glucosamine 6-phosphate (according to Swiss-Prot)
- Protein family: phosphohexose mutase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU01770
- UniProt: O34824
- KEGG entry: [2]
- E.C. number: 5.4.2.10
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- pGP400: (expression in B. subtilis in pBQ200), available in Jörg Stülke's lab
- pGP1401: (expression, purification in E. coli with N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- pGP1402: (cloning vector for glmM (GlmM S100A) in E. coli, in pBlueskript KS), available in Jörg Stülke's lab
- pGP1403: (expression of GlmM (S100A) in B. subtilis in pBQ200), available in Jörg Stülke's lab
- pGP1405: (expression, purification of GlmM (S100A) in E. coli with N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- lacZ fusion:
- GP1340 (cat) based on pAC6, available in Jörg Stülke's lab
- GFP fusion:
- GP1382 glmM-gfp ermC (based on pGP1080), available in Jörg Stülke's lab
- two-hybrid system:
- pGP2576 (glmM in p25N), available in Jörg Stülke's lab
- FLAG-tag construct:
- GP1380 lacA::cdaR-Strep aphA3 glmM-3xFLAG ermC (based on pGP1460 and pGP1087), available in Jörg Stülke's lab
- Strep-tag construct:
- GP1388 lacA::glmM-Strep aphA3 cdaA-3xFLAG ermC (based on pGP1460 and pGP1087), available in Jörg Stülke's lab
- Antibody: available in Jörg Stülke's lab
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Felix M P Mehne, Katrin Gunka, Hinnerk Eilers, Christina Herzberg, Volkhard Kaever, Jörg Stülke
Cyclic di-AMP homeostasis in bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth.
J Biol Chem: 2013, 288(3);2004-17
[PubMed:23192352]
[WorldCat.org]
[DOI]
(I p)
Ritcha Mehra-Chaudhary, Jacob Mick, Lesa J Beamer
Crystal structure of Bacillus anthracis phosphoglucosamine mutase, an enzyme in the peptidoglycan biosynthetic pathway.
J Bacteriol: 2011, 193(16);4081-7
[PubMed:21685296]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Sebastian R Schmidl, Katrin Gronau, Nico Pietack, Michael Hecker, Dörte Becher, Jörg Stülke
The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases.
Mol Cell Proteomics: 2010, 9(6);1228-42
[PubMed:20097688]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
L Jolly, F Pompeo, J van Heijenoort, F Fassy, D Mengin-Lecreulx
Autophosphorylation of phosphoglucosamine mutase from Escherichia coli.
J Bacteriol: 2000, 182(5);1280-5
[PubMed:10671448]
[WorldCat.org]
[DOI]
(P p)
L Jolly, P Ferrari, D Blanot, J Van Heijenoort, F Fassy, D Mengin-Lecreulx
Reaction mechanism of phosphoglucosamine mutase from Escherichia coli.
Eur J Biochem: 1999, 262(1);202-10
[PubMed:10231382]
[WorldCat.org]
[DOI]
(P p)
D Mengin-Lecreulx, J van Heijenoort
Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli.
J Biol Chem: 1996, 271(1);32-9
[PubMed:8550580]
[WorldCat.org]
[DOI]
(P p)