Difference between revisions of "Ggt"
(→References) |
(→References) |
||
| Line 136: | Line 136: | ||
=References= | =References= | ||
| − | <pubmed>15583164,12892879,18957862,20088880 22383849 23335395,23033921,21513304,21987357 24279353 </pubmed> | + | <pubmed>15583164,12892879,18957862,20088880 22383849 23335395,23033921,21513304,21987357 24279353 24531494 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:19, 18 February 2014
- Description: gamma-glutamyltransferase
| Gene name | ggt |
| Synonyms | pac |
| Essential | no |
| Product | gamma-glutamyltransferase |
| Function | degradation of poly-glutamate capsules |
| Gene expression levels in SubtiExpress: ggt | |
| MW, pI | 64 kDa, 5.453 |
| Gene length, protein length | 1761 bp, 587 aa |
| Immediate neighbours | yoeD, yofA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 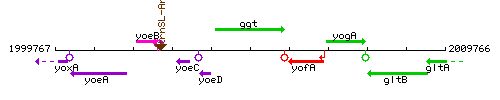
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed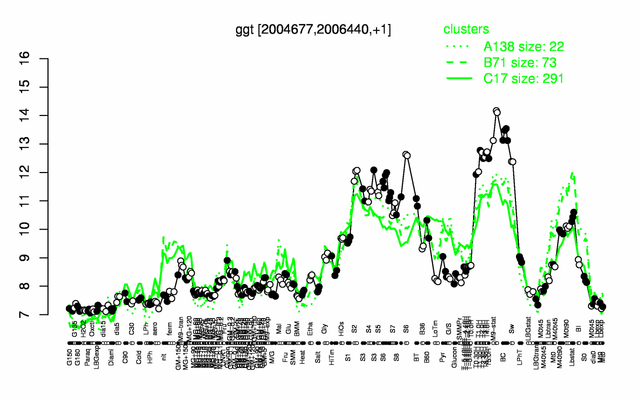
| |
Contents
Categories containing this gene/protein
capsule biosynthesis and degradation
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU18410
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (5-L-glutamyl)-peptide + an amino acid = peptide + 5-L-glutamyl amino acid (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- extracellular (signal peptide) PubMed
Database entries
- Structure: 2V36
- UniProt: P54422
- KEGG entry: [2]
- E.C. number: 2.3.2.2
Additional information
Expression and regulation
- Regulation:
- strongly induced in response to glucose starvation in M9 medium PubMed
- Regulatory mechanism:
- Additional information:
- translation is likely to require Efp due to the presence of several consecutive proline residues PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Tomoyo Ida, Hideyuki Suzuki, Keiichi Fukuyama, Jun Hiratake, Kei Wada
Structure of Bacillus subtilis γ-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.
Acta Crystallogr D Biol Crystallogr: 2014, 70(Pt 2);607-14
[PubMed:24531494]
[WorldCat.org]
[DOI]
(I p)
Carlo F Morelli, Cinzia Calvio, Marco Biagiotti, Giovanna Speranza
pH-dependent hydrolase, glutaminase, transpeptidase and autotranspeptidase activities of Bacillus subtilis γ-glutamyltransferase.
FEBS J: 2014, 281(1);232-45
[PubMed:24279353]
[WorldCat.org]
[DOI]
(I p)
Viola Scoffone, Daniele Dondi, Ginevra Biino, Giovanni Borghese, Dario Pasini, Alessandro Galizzi, Cinzia Calvio
Knockout of pgdS and ggt genes improves γ-PGA yield in B. subtilis.
Biotechnol Bioeng: 2013, 110(7);2006-12
[PubMed:23335395]
[WorldCat.org]
[DOI]
(I p)
Imke G de Jong, Jan-Willem Veening, Oscar P Kuipers
Single cell analysis of gene expression patterns during carbon starvation in Bacillus subtilis reveals large phenotypic variation.
Environ Microbiol: 2012, 14(12);3110-21
[PubMed:23033921]
[WorldCat.org]
[DOI]
(I p)
Pierre Nicolas, Ulrike Mäder, Etienne Dervyn, Tatiana Rochat, Aurélie Leduc, Nathalie Pigeonneau, Elena Bidnenko, Elodie Marchadier, Mark Hoebeke, Stéphane Aymerich, Dörte Becher, Paola Bisicchia, Eric Botella, Olivier Delumeau, Geoff Doherty, Emma L Denham, Mark J Fogg, Vincent Fromion, Anne Goelzer, Annette Hansen, Elisabeth Härtig, Colin R Harwood, Georg Homuth, Hanne Jarmer, Matthieu Jules, Edda Klipp, Ludovic Le Chat, François Lecointe, Peter Lewis, Wolfram Liebermeister, Anika March, Ruben A T Mars, Priyanka Nannapaneni, David Noone, Susanne Pohl, Bernd Rinn, Frank Rügheimer, Praveen K Sappa, Franck Samson, Marc Schaffer, Benno Schwikowski, Leif Steil, Jörg Stülke, Thomas Wiegert, Kevin M Devine, Anthony J Wilkinson, Jan Maarten van Dijl, Michael Hecker, Uwe Völker, Philippe Bessières, Philippe Noirot
Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis.
Science: 2012, 335(6072);1103-6
[PubMed:22383849]
[WorldCat.org]
[DOI]
(I p)
Weiqiao Li, Bo Jiang, Wanmeng Mu, Ming Miao, Tao Zhang
Effects of pH and dissolved oxygen on the synthesis of γ-glutamyltranspeptidase from Bacillus subtilis SK 11.004.
J Sci Food Agric: 2012, 92(3);475-80
[PubMed:21987357]
[WorldCat.org]
[DOI]
(I p)
Yuying Shuai, Tao Zhang, Wanmeng Mu, Bo Jiang
Purification and characterization of γ-glutamyltranspeptidase from Bacillus subtilis SK11.004.
J Agric Food Chem: 2011, 59(11);6233-8
[PubMed:21513304]
[WorldCat.org]
[DOI]
(I p)
Kei Wada, Machiko Irie, Hideyuki Suzuki, Keiichi Fukuyama
Crystal structure of the halotolerant gamma-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket.
FEBS J: 2010, 277(4);1000-9
[PubMed:20088880]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Keitarou Kimura, Lam-Son Phan Tran, Ikuo Uchida, Yoshifumi Itoh
Characterization of Bacillus subtilis gamma-glutamyltransferase and its involvement in the degradation of capsule poly-gamma-glutamate.
Microbiology (Reading): 2004, 150(Pt 12);4115-23
[PubMed:15583164]
[WorldCat.org]
[DOI]
(P p)
Hiromichi Minami, Hideyuki Suzuki, Hidehiko Kumagai
A mutant Bacillus subtilis gamma-glutamyltranspeptidase specialized in hydrolysis activity.
FEMS Microbiol Lett: 2003, 224(2);169-73
[PubMed:12892879]
[WorldCat.org]
[DOI]
(P p)