Difference between revisions of "GcvT"
(→References) |
|||
| Line 142: | Line 142: | ||
=References= | =References= | ||
| − | <pubmed>,15096624,15472076, 23249744 </pubmed> | + | <pubmed>,15096624,15472076, 23249744 23721735 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:15, 4 June 2013
- Description: aminomethyltransferase (glycine cleavage system protein T)
| Gene name | gcvT |
| Synonyms | yqhI |
| Essential | no |
| Product | aminomethyltransferase (glycine cleavage system protein T) |
| Function | glycine utilization |
| Gene expression levels in SubtiExpress: gcvT | |
| MW, pI | 39 kDa, 5.734 |
| Gene length, protein length | 1086 bp, 362 aa |
| Immediate neighbours | gcvPA, yqhH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 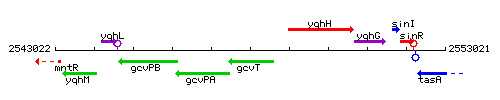
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed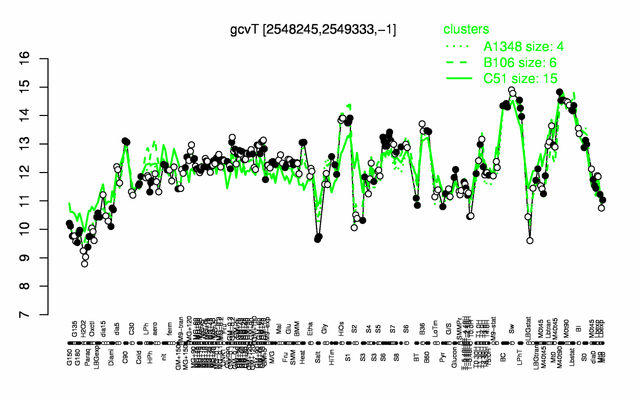
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU24570
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: [Protein]-S(8)-aminomethyldihydrolipoyllysine + tetrahydrofolate = [protein]-dihydrolipoyllysine + 5,10-methylenetetrahydrofolate + NH3 (according to Swiss-Prot)
- Protein family: gcvT family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1YX2
- UniProt: P54378
- KEGG entry: [3]
- E.C. number: 2.1.2.10
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- induced by glycine PubMed
- Regulatory mechanism:
- induction requires a co-operatively acting riboswitch (Gly-box) PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Kokoro Hamachi, Hikari Hayashi, Miyuki Shimamura, Yuiha Yamaji, Ai Kaneko, Aruma Fujisawa, Takuya Umehara, Koji Tamura
Glycols modulate terminator stem stability and ligand-dependency of a glycine riboswitch.
Biosystems: 2013, 113(2);59-65
[PubMed:23721735]
[WorldCat.org]
[DOI]
(I p)
Nathan J Baird, Adrian R Ferré-D'Amaré
Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches.
RNA: 2013, 19(2);167-76
[PubMed:23249744]
[WorldCat.org]
[DOI]
(I p)
Maumita Mandal, Mark Lee, Jeffrey E Barrick, Zasha Weinberg, Gail Mitchell Emilsson, Walter L Ruzzo, Ronald R Breaker
A glycine-dependent riboswitch that uses cooperative binding to control gene expression.
Science: 2004, 306(5694);275-9
[PubMed:15472076]
[WorldCat.org]
[DOI]
(I p)
Jeffrey E Barrick, Keith A Corbino, Wade C Winkler, Ali Nahvi, Maumita Mandal, Jennifer Collins, Mark Lee, Adam Roth, Narasimhan Sudarsan, Inbal Jona, J Kenneth Wickiser, Ronald R Breaker
New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control.
Proc Natl Acad Sci U S A: 2004, 101(17);6421-6
[PubMed:15096624]
[WorldCat.org]
[DOI]
(P p)