Difference between revisions of "GabR"
(→References) |
|||
| Line 138: | Line 138: | ||
=References= | =References= | ||
| − | <pubmed>15223311,12123465, 24127574 22020104 </pubmed> | + | <pubmed> 11756427 15223311,12123465, 24127574 22020104 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:16, 22 November 2013
- Description: transcription activator of gabT-gabD, repressor of gabR (MocR/ GabR family)
| Gene name | gabR |
| Synonyms | ycnF |
| Essential | no |
| Product | transcription regulator |
| Function | regulation of gamma-amino butyric acid utilization |
| Gene expression levels in SubtiExpress: gabR | |
| MW, pI | 54 kDa, 6.709 |
| Gene length, protein length | 1437 bp, 479 aa |
| Immediate neighbours | yczG, gabT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 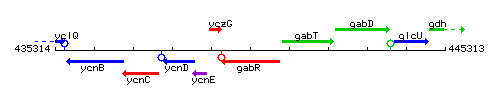
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
utilization of amino acids, transcription factors and their control
This gene is a member of the following regulons
The GabR regulon:
The gene
Basic information
- Locus tag: BSU03890
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: MocR/ GabR family PubMed
- Paralogous protein(s): none, but there are 7 members of the MocR/ GabR family in B. subtilis
Extended information on the protein
- Kinetic information:
- Domains: N-terminal DNA-binding helix-turn-helix motif; C-terminal domain is homologous to PLP-binding large domain of aminotransferases.
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P94426
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- GabR: negative autoregulation PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Your additional remarks
References
Raji Edayathumangalam, Rui Wu, Roman Garcia, Yuguang Wang, Wei Wang, Cheryl A Kreinbring, Alicia Bach, Jingling Liao, Todd A Stone, Thomas C Terwilliger, Quyen Q Hoang, Boris R Belitsky, Gregory A Petsko, Dagmar Ringe, Dali Liu
Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc Natl Acad Sci U S A: 2013, 110(44);17820-5
[PubMed:24127574]
[WorldCat.org]
[DOI]
(I p)
E Bramucci, T Milano, S Pascarella
Genomic distribution and heterogeneity of MocR-like transcriptional factors containing a domain belonging to the superfamily of the pyridoxal-5'-phosphate dependent enzymes of fold type I.
Biochem Biophys Res Commun: 2011, 415(1);88-93
[PubMed:22020104]
[WorldCat.org]
[DOI]
(I p)
Boris R Belitsky
Bacillus subtilis GabR, a protein with DNA-binding and aminotransferase domains, is a PLP-dependent transcriptional regulator.
J Mol Biol: 2004, 340(4);655-64
[PubMed:15223311]
[WorldCat.org]
[DOI]
(P p)
Boris R Belitsky, Abraham L Sonenshein
GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis.
Mol Microbiol: 2002, 45(2);569-83
[PubMed:12123465]
[WorldCat.org]
[DOI]
(P p)
Sébastien Rigali, Adeline Derouaux, Fabrizio Giannotta, Jean Dusart
Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies.
J Biol Chem: 2002, 277(15);12507-15
[PubMed:11756427]
[WorldCat.org]
[DOI]
(P p)