FusA
- Description: elongation factor G, facilitates movement of tRNA–mRNA by one codon
| Gene name | fusA |
| Synonyms | fus |
| Essential | yes PubMed |
| Product | elongation factor G |
| Function | translation |
| Gene expression levels in SubtiExpress: fusA | |
| Interactions involving this protein in SubtInteract: FusA | |
| MW, pI | 76 kDa, 4.615 |
| Gene length, protein length | 2076 bp, 692 aa |
| Immediate neighbours | rpsG, tufA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed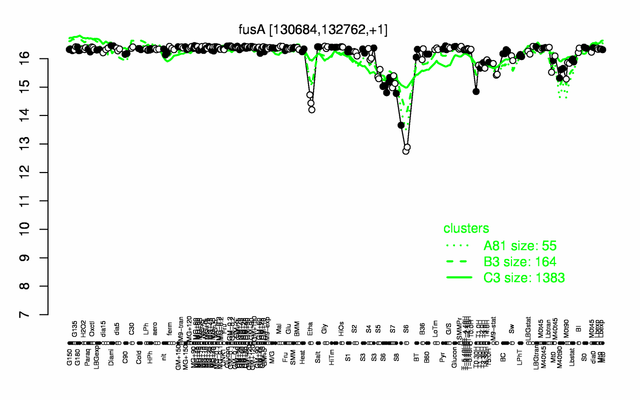
| |
Contents
Categories containing this gene/protein
translation, essential genes, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01120
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU01120
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: hydrolyses GTP
- Protein family: EF-G/EF-2 subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylation on Ser-213 AND Ser-302 AND Ser-569 AND Ser-680 AND (Thr-24 OR Thr-25) AND (Thr-43 OR Ser 48) PubMed, PubMed
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU01120
- Structure:
- UniProt: P80868
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 11452 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 49524 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 23259 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 12858 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 16612 PubMed
Biological materials
- Mutant:
- Expression vector:
- for expression/ purification from B. subtilis with N-terminal Strep-tag, for SPINE, in pGP380: pGP840, available in Jörg Stülke's lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP848, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yun Chen, Shu Feng, Veerendra Kumar, Rya Ero, Yong-Gui Gao
Structure of EF-G-ribosome complex in a pretranslocation state.
Nat Struct Mol Biol: 2013, 20(9);1077-84
[PubMed:23912278]
[WorldCat.org]
[DOI]
(I p)
Nina Clementi, Anna Chirkova, Barbara Puffer, Ronald Micura, Norbert Polacek
Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation.
Nat Chem Biol: 2010, 6(5);344-51
[PubMed:20348921]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
A AEvarsson, E Brazhnikov, M Garber, J Zheltonosova, Y Chirgadze, S al-Karadaghi, L A Svensson, A Liljas
Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus.
EMBO J: 1994, 13(16);3669-77
[PubMed:8070397]
[WorldCat.org]
[DOI]
(P p)