Difference between revisions of "FlgD"
(→Expression and regulation) |
(→Expression and regulation) |
||
| Line 107: | Line 107: | ||
** ''[[ylxF]]-[[fliK]]-[[flgD]]-[[flgE]]-[[fliL]]-[[fliM]]-[[fliY]]-[[cheY]]-[[fliZ]]-[[fliP]]-[[fliQ]]-[[fliR]]-[[flhB]]-[[flhA]]-[[flhF]]-[[ylxH]]-[[cheB]]-[[cheA]]-[[cheW]]-[[cheC]]-[[cheD]]-[[sigD]]-[[swrB]]'' {{PubMed|20233303}} | ** ''[[ylxF]]-[[fliK]]-[[flgD]]-[[flgE]]-[[fliL]]-[[fliM]]-[[fliY]]-[[cheY]]-[[fliZ]]-[[fliP]]-[[fliQ]]-[[fliR]]-[[flhB]]-[[flhA]]-[[flhF]]-[[ylxH]]-[[cheB]]-[[cheA]]-[[cheW]]-[[cheC]]-[[cheD]]-[[sigD]]-[[swrB]]'' {{PubMed|20233303}} | ||
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=flgD_1699738_1700160_1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=flgD_1699738_1700160_1 flgD] {{PubMed|22383849}} |
* '''Sigma factor:''' | * '''Sigma factor:''' | ||
Revision as of 13:02, 27 June 2012
- Description: putative flagellar hook cap, required for hook assembly
| Gene name | flgD |
| Synonyms | ylxG |
| Essential | no |
| Product | putative flagellar hook cap |
| Function | flagellar hook assembly |
| MW, pI | 15 kDa, 4.351 |
| Gene length, protein length | 420 bp, 140 aa |
| Immediate neighbours | fliK, flgE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 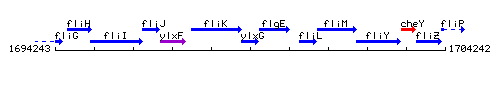
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed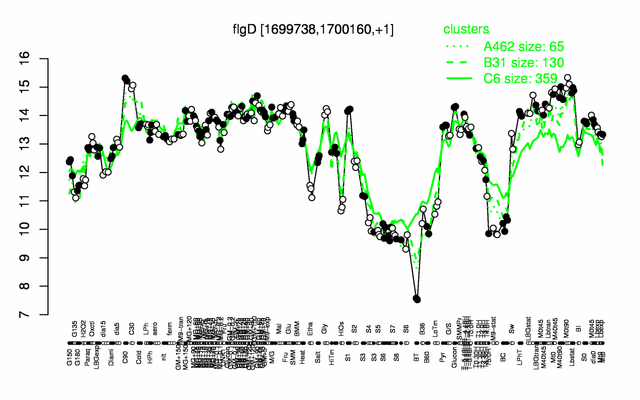
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
CodY regulon, SigD regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU16280
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: flagellar hook assembly PubMed
- Protein family: flgD family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P23455
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Kazuo Kobayashi
Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2007, 66(2);395-409
[PubMed:17850253]
[WorldCat.org]
[DOI]
(P p)
H Werhane, P Lopez, M Mendel, M Zimmer, G W Ordal, L M Márquez-Magaña
The last gene of the fla/che operon in Bacillus subtilis, ylxL, is required for maximal sigmaD function.
J Bacteriol: 2004, 186(12);4025-9
[PubMed:15175317]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
W Estacio, S S Anna-Arriola, M Adedipe, L M Márquez-Magaña
Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis.
J Bacteriol: 1998, 180(14);3548-55
[PubMed:9657996]
[WorldCat.org]
[DOI]
(P p)
L M Márquez-Magaña, M J Chamberlin
Characterization of the sigD transcription unit of Bacillus subtilis.
J Bacteriol: 1994, 176(8);2427-34
[PubMed:8157612]
[WorldCat.org]
[DOI]
(P p)