Difference between revisions of "FbpB"
| Line 1: | Line 1: | ||
| − | + | ||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || RNA chaperone | |style="background:#ABCDEF;" align="center"|'''Function''' || RNA chaperone | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU04530 fbpB] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 6 kDa, 9.9 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 6 kDa, 9.9 | ||
Revision as of 15:16, 6 August 2012
| Gene name | fbpB |
| Synonyms | ydbN |
| Essential | no |
| Product | Fur-regulated basic protein, acts as RNA chaperone for fsrA, response to iron limitation |
| Function | RNA chaperone |
| Gene expression levels in SubtiExpress: fbpB | |
| MW, pI | 6 kDa, 9.9 |
| Gene length, protein length | 177 bp, 59 aa |
| Immediate neighbours | ydbM, fbpA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 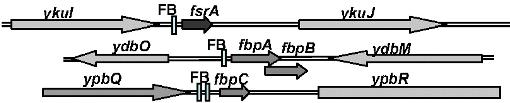
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
acquisition of iron, iron metabolism, RNA chaperones, short peptides
This gene is a member of the following regulons
The FbpB regulon: lutA-lutB-lutC
The gene
Basic information
- Locus tag: BSU04530
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The sequence of the former ydbN gene was corrected, and does now contain two genes, fbpA and fbpB. PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P96609
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Gregory T Smaldone, Haike Antelmann, Ahmed Gaballa, John D Helmann
The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases.
J Bacteriol: 2012, 194(10);2586-93
[PubMed:22427629]
[WorldCat.org]
[DOI]
(I p)
Gregory T Smaldone, Olga Revelles, Ahmed Gaballa, Uwe Sauer, Haike Antelmann, John D Helmann
A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism.
J Bacteriol: 2012, 194(10);2594-605
[PubMed:22389480]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Ahmed Gaballa, Haike Antelmann, Claudio Aguilar, Sukhjit K Khakh, Kyung-Bok Song, Gregory T Smaldone, John D Helmann
The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins.
Proc Natl Acad Sci U S A: 2008, 105(33);11927-32
[PubMed:18697947]
[WorldCat.org]
[DOI]
(I p)
Noel Baichoo, Tao Wang, Rick Ye, John D Helmann
Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon.
Mol Microbiol: 2002, 45(6);1613-29
[PubMed:12354229]
[WorldCat.org]
[DOI]
(P p)