Difference between revisions of "FapR"
| (49 intermediate revisions by 8 users not shown) | |||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of fatty acid biosynthesis | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of fatty acid biosynthesis | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU15880 fapR] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=FapR FapR] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=fapR fapR]''' | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 5.393 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 5.393 | ||
| Line 20: | Line 26: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[recG]]'', ''[[plsX]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[recG]]'', ''[[plsX]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU15880 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU15880 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU15880 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ylpC_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ylpC_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=fapR_1661967_1662533_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:fapR_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU15880]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/> | ||
| − | + | = [[Categories]] containing this gene/protein = | |
| + | {{SubtiWiki category|[[biosynthesis of lipids]]}}, | ||
| + | {{SubtiWiki category|[[transcription factors and their control]]}} | ||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[ComA regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[FapR regulon]]}} | ||
| + | =The [[FapR regulon]]= | ||
=The gene= | =The gene= | ||
=== Basic information === | === Basic information === | ||
| − | * ''' | + | * '''Locus tag:''' BSU15880 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15880&redirect=T BSU15880] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/fapR-plsX-fabDG.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/fapR-plsX-fabDG.html] | ||
| Line 46: | Line 65: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
=The protein= | =The protein= | ||
| Line 52: | Line 70: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' FapR regulates the expression of at least 10 genes (the ''fap'' regulon). It autoregulates its own expression. Malonyl-CoA, a precursor of fatty acid biosynthesis, binds to FapR changing its conformation to a non-DNA binding state. Hence, conditions that cause malonyl-CoA accumulation, like fatty acid biosynthesis inhibition, derepress the ''fap'' regulon. |
| − | * '''Protein family:''' | + | * '''Protein family:''' fapR family (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 68: | Line 86: | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' malonyl-CoA and malonyl-[[AcpA|ACP]] act as the molecular inducer of the FapR regulon {{PubMed|20201588}} |
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** malonyl-[[AcpA|ACP]]-[[FapR]] {{PubMed|20201588}} | ||
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15880&redirect=T BSU15880] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2F3X 2F3X] (complex with effector), [http://www.rcsb.org/pdb/explore.do?structureId=2F41 2F41] | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2F3X 2F3X] (complex with effector), [http://www.rcsb.org/pdb/explore.do?structureId=2F41 2F41] | ||
| − | * ''' | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/O34835 O34835] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU15880] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 108: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[fapR]]-[[plsX]]-[[fabD]]-[[fabG]]-[[acpA]]'' {{PubMed|12737802,19850612}} |
| + | |||
| + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=fapR_1661967_1662533_1 fapR] {{PubMed|22383849}} | ||
| − | * ''' | + | * '''Sigma factor:''' [[SigA]] {{PubMed|16932747}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** repressed in the absence of malonyl-CoA or malonyl-[[AcpA|ACP]] ([[FapR]]) {{PubMed|20201588}} | ||
| + | ** expressed at high cell density ([[ComA]]) {{PubMed|16091051}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[ComA]]: transcription activation ([[ComA]]) {{PubMed|16091051}} | ||
| + | ** [[FapR]]: transcription repression {{PubMed|12737802}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 85 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 490 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 412 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 729 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
| Line 117: | Line 146: | ||
=References= | =References= | ||
| + | ==Reviews== | ||
| + | <pubmed>17919287,</pubmed> | ||
| + | ==The [[FapR regulon]]== | ||
| + | <pubmed>12737802, </pubmed> | ||
| + | ==Other original publications== | ||
| + | '''Additional publications:''' {{PubMed|23102226}} | ||
| + | <pubmed>18941141, 18820022, 18384517, 16932747,19850612 16091051 20201588 </pubmed> | ||
| − | + | [[Category:Protein-coding genes]] | |
Latest revision as of 14:19, 17 April 2014
- Description: repressor of fatty acid synthetic genes
| Gene name | fapR |
| Synonyms | ylpC |
| Essential | no |
| Product | transcriptional repressor |
| Function | regulation of fatty acid biosynthesis |
| Gene expression levels in SubtiExpress: fapR | |
| Interactions involving this protein in SubtInteract: FapR | |
| Metabolic function and regulation of this protein in SubtiPathways: fapR | |
| MW, pI | 21 kDa, 5.393 |
| Gene length, protein length | 564 bp, 188 aa |
| Immediate neighbours | recG, plsX |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed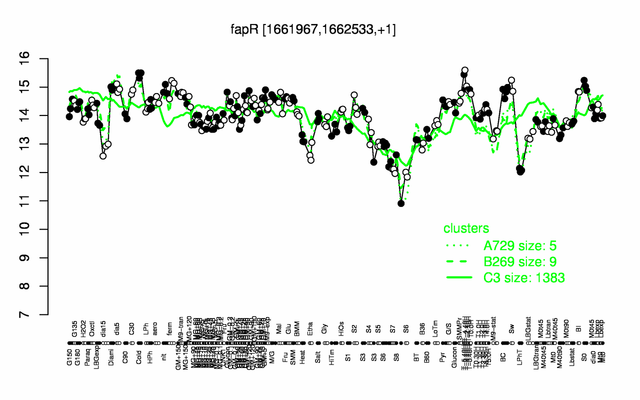
| |
Contents
Categories containing this gene/protein
biosynthesis of lipids, transcription factors and their control
This gene is a member of the following regulons
The FapR regulon
The gene
Basic information
- Locus tag: BSU15880
Phenotypes of a mutant
Database entries
- BsubCyc: BSU15880
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: FapR regulates the expression of at least 10 genes (the fap regulon). It autoregulates its own expression. Malonyl-CoA, a precursor of fatty acid biosynthesis, binds to FapR changing its conformation to a non-DNA binding state. Hence, conditions that cause malonyl-CoA accumulation, like fatty acid biosynthesis inhibition, derepress the fap regulon.
- Protein family: fapR family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: malonyl-CoA and malonyl-ACP act as the molecular inducer of the FapR regulon PubMed
Database entries
- BsubCyc: BSU15880
- UniProt: O34835
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 85 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 490 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 412 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 729 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Yasutaro Fujita, Hiroshi Matsuoka, Kazutake Hirooka
Regulation of fatty acid metabolism in bacteria.
Mol Microbiol: 2007, 66(4);829-39
[PubMed:17919287]
[WorldCat.org]
[DOI]
(P p)
The FapR regulon
Gustavo E Schujman, Luciana Paoletti, Alan D Grossman, Diego de Mendoza
FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis.
Dev Cell: 2003, 4(5);663-72
[PubMed:12737802]
[WorldCat.org]
[DOI]
(P p)
Other original publications
Additional publications: PubMed
Mariano A Martinez, María-Eugenia Zaballa, Francis Schaeffer, Marco Bellinzoni, Daniela Albanesi, Gustavo E Schujman, Alejandro J Vila, Pedro M Alzari, Diego de Mendoza
A novel role of malonyl-ACP in lipid homeostasis.
Biochemistry: 2010, 49(14);3161-7
[PubMed:20201588]
[WorldCat.org]
[DOI]
(I p)
Mariano A Martinez, Diego de Mendoza, Gustavo E Schujman
Transcriptional and functional characterization of the gene encoding acyl carrier protein in Bacillus subtilis.
Microbiology (Reading): 2010, 156(Pt 2);484-495
[PubMed:19850612]
[WorldCat.org]
[DOI]
(I p)
Yong-Mei Zhang, Charles O Rock
Transcriptional regulation in bacterial membrane lipid synthesis.
J Lipid Res: 2009, 50 Suppl(Suppl);S115-9
[PubMed:18941141]
[WorldCat.org]
[DOI]
(P p)
Letal I Salzberg, John D Helmann
Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition.
J Bacteriol: 2008, 190(23);7797-807
[PubMed:18820022]
[WorldCat.org]
[DOI]
(I p)
Gustavo E Schujman, Silvia Altabe, Diego de Mendoza
A malonyl-CoA-dependent switch in the bacterial response to a dysfunction of lipid metabolism.
Mol Microbiol: 2008, 68(4);987-96
[PubMed:18384517]
[WorldCat.org]
[DOI]
(I p)
Gustavo E Schujman, Marcelo Guerin, Alejandro Buschiazzo, Francis Schaeffer, Leticia I Llarrull, Georgina Reh, Alejandro J Vila, Pedro M Alzari, Diego de Mendoza
Structural basis of lipid biosynthesis regulation in Gram-positive bacteria.
EMBO J: 2006, 25(17);4074-83
[PubMed:16932747]
[WorldCat.org]
[DOI]
(P p)
Natalia Comella, Alan D Grossman
Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis.
Mol Microbiol: 2005, 57(4);1159-74
[PubMed:16091051]
[WorldCat.org]
[DOI]
(P p)