Difference between revisions of "FabF"
(→Database entries) |
|||
| Line 96: | Line 96: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?structureId= | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?structureId=4LS5 4LS5] {{PubMed|24641521}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O34340 O34340] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O34340 O34340] | ||
Revision as of 09:46, 20 March 2014
- Description: beta-ketoacyl-acyl carrier protein synthase II, involved in the control of membrane fluidity
| Gene name | fabF |
| Synonyms | yjaY |
| Essential | yes PubMed |
| Product | beta-ketoacyl-acyl carrier protein synthase II |
| Function | fatty acid biosynthesis |
| Gene expression levels in SubtiExpress: fabF | |
| Metabolic function and regulation of this protein in SubtiPathways: fabF | |
| MW, pI | 43 kDa, 4.768 |
| Gene length, protein length | 1239 bp, 413 aa |
| Immediate neighbours | fabHA, yjaZ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 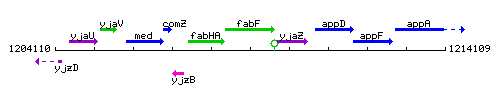
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed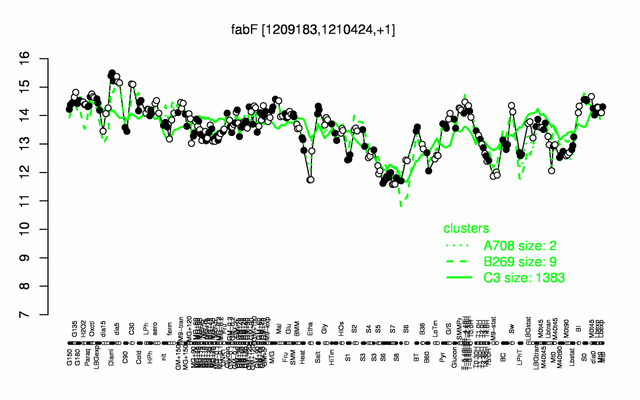
| |
Contents
Categories containing this gene/protein
membrane dynamics, biosynthesis of lipids, cell envelope stress proteins (controlled by SigM, V, W, X, Y), essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU11340
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: beta-ketoacyl-ACP synthases family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- UniProt: O34340
- KEGG entry: [3]
- E.C. number: 2.3.1.179
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Yasutaro Fujita, Hiroshi Matsuoka, Kazutake Hirooka
Regulation of fatty acid metabolism in bacteria.
Mol Microbiol: 2007, 66(4);829-39
[PubMed:17919287]
[WorldCat.org]
[DOI]
(P p)
Stephen W White, Jie Zheng, Yong-Mei Zhang, Rock
The structural biology of type II fatty acid biosynthesis.
Annu Rev Biochem: 2005, 74;791-831
[PubMed:15952903]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Felipe Trajtenberg, Silvia Altabe, Nicole Larrieux, Florencia Ficarra, Diego de Mendoza, Alejandro Buschiazzo, Gustavo E Schujman
Structural insights into bacterial resistance to cerulenin.
FEBS J: 2014, 281(10);2324-38
[PubMed:24641521]
[WorldCat.org]
[DOI]
(I p)
Yong Heon Lee, Anthony W Kingston, John D Helmann
Glutamate dehydrogenase affects resistance to cell wall antibiotics in Bacillus subtilis.
J Bacteriol: 2012, 194(5);993-1001
[PubMed:22178969]
[WorldCat.org]
[DOI]
(I p)
Anthony W Kingston, Chitra Subramanian, Charles O Rock, John D Helmann
A σW-dependent stress response in Bacillus subtilis that reduces membrane fluidity.
Mol Microbiol: 2011, 81(1);69-79
[PubMed:21542858]
[WorldCat.org]
[DOI]
(I p)
Michaela Wenzel, Malay Patra, Dirk Albrecht, David Y-K Chen, K C Nicolaou, Nils Metzler-Nolte, Julia E Bandow
Proteomic signature of fatty acid biosynthesis inhibition available for in vivo mechanism-of-action studies.
Antimicrob Agents Chemother: 2011, 55(6);2590-6
[PubMed:21383089]
[WorldCat.org]
[DOI]
(I p)
Gustavo E Schujman, Silvia Altabe, Diego de Mendoza
A malonyl-CoA-dependent switch in the bacterial response to a dysfunction of lipid metabolism.
Mol Microbiol: 2008, 68(4);987-96
[PubMed:18384517]
[WorldCat.org]
[DOI]
(I p)
Allen C Price, Charles O Rock, Stephen W White
The 1.3-Angstrom-resolution crystal structure of beta-ketoacyl-acyl carrier protein synthase II from Streptococcus pneumoniae.
J Bacteriol: 2003, 185(14);4136-43
[PubMed:12837788]
[WorldCat.org]
[DOI]
(P p)
Gustavo E Schujman, Luciana Paoletti, Alan D Grossman, Diego de Mendoza
FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis.
Dev Cell: 2003, 4(5);663-72
[PubMed:12737802]
[WorldCat.org]
[DOI]
(P p)
K Kobayashi, S D Ehrlich, A Albertini, G Amati, K K Andersen, M Arnaud, K Asai, S Ashikaga, S Aymerich, P Bessieres, F Boland, S C Brignell, S Bron, K Bunai, J Chapuis, L C Christiansen, A Danchin, M Débarbouille, E Dervyn, E Deuerling, K Devine, S K Devine, O Dreesen, J Errington, S Fillinger, S J Foster, Y Fujita, A Galizzi, R Gardan, C Eschevins, T Fukushima, K Haga, C R Harwood, M Hecker, D Hosoya, M F Hullo, H Kakeshita, D Karamata, Y Kasahara, F Kawamura, K Koga, P Koski, R Kuwana, D Imamura, M Ishimaru, S Ishikawa, I Ishio, D Le Coq, A Masson, C Mauël, R Meima, R P Mellado, A Moir, S Moriya, E Nagakawa, H Nanamiya, S Nakai, P Nygaard, M Ogura, T Ohanan, M O'Reilly, M O'Rourke, Z Pragai, H M Pooley, G Rapoport, J P Rawlins, L A Rivas, C Rivolta, A Sadaie, Y Sadaie, M Sarvas, T Sato, H H Saxild, E Scanlan, W Schumann, J F M L Seegers, J Sekiguchi, A Sekowska, S J Séror, M Simon, P Stragier, R Studer, H Takamatsu, T Tanaka, M Takeuchi, H B Thomaides, V Vagner, J M van Dijl, K Watabe, A Wipat, H Yamamoto, M Yamamoto, Y Yamamoto, K Yamane, K Yata, K Yoshida, H Yoshikawa, U Zuber, N Ogasawara
Essential Bacillus subtilis genes.
Proc Natl Acad Sci U S A: 2003, 100(8);4678-83
[PubMed:12682299]
[WorldCat.org]
[DOI]
(P p)