Difference between revisions of "ExoA"
(→Extended information on the protein) |
|||
| Line 97: | Line 97: | ||
* '''Operon:''' ''[[exoA]]-[[ccpB]]'' (according to [http://dbtbs.hgc.jp/COG/prom/exoA-ccpA.html DBTBS]) | * '''Operon:''' ''[[exoA]]-[[ccpB]]'' (according to [http://dbtbs.hgc.jp/COG/prom/exoA-ccpA.html DBTBS]) | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=exoAA_4197780_4198538_-1 exoA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigG]] {{PubMed|16237020}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 104: | Line 106: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 10:38, 17 April 2012
- Description: apurinic/apyrimidinic endonuclease, multifunctional DNA-repair enzyme, important for spore dormance
| Gene name | exoA |
| Synonyms | |
| Essential | no |
| Product | apurinic/apyrimidinic endonuclease |
| Function | repair of oxidative DNA damage in spores |
| MW, pI | 29 kDa, 4.592 |
| Gene length, protein length | 756 bp, 252 aa |
| Immediate neighbours | ccpB, rpsR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 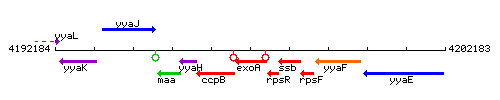
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40880
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Exonucleolytic cleavage in the 3'- to 5'-direction to yield nucleoside 5'-phosphates (according to Swiss-Prot)
- Protein family: DNA repair enzymes AP/exoA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P37454
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- available in Mario Pedraza-Reyes' lab
- GP898 (exoA::kan), available in Stülke lab
- GP1503 (exoA::kan, nfo::cat), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Marcelo Barraza-Salas, Juan R Ibarra-Rodríguez, Silvia J Mellado, José M Salas-Pacheco, Peter Setlow, Mario Pedraza-Reyes
Effects of forespore-specific overexpression of apurinic/apyrimidinic endonuclease Nfo on the DNA-damage resistance properties of Bacillus subtilis spores.
FEMS Microbiol Lett: 2010, 302(2);159-65
[PubMed:19930460]
[WorldCat.org]
[DOI]
(I p)
Juan R Ibarra, Alma D Orozco, Juan A Rojas, Karina López, Peter Setlow, Ronald E Yasbin, Mario Pedraza-Reyes
Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores.
J Bacteriol: 2008, 190(6);2031-8
[PubMed:18203828]
[WorldCat.org]
[DOI]
(I p)
José M Salas-Pacheco, Barbara Setlow, Peter Setlow, Mario Pedraza-Reyes
Role of the Nfo (YqfS) and ExoA apurinic/apyrimidinic endonucleases in protecting Bacillus subtilis spores from DNA damage.
J Bacteriol: 2005, 187(21);7374-81
[PubMed:16237020]
[WorldCat.org]
[DOI]
(P p)
T Shida, T Ogawa, N Ogasawara, J Sekiguchi
Characterization of Bacillus subtilis ExoA protein: a multifunctional DNA-repair enzyme similar to the Escherichia coli exonuclease III.
Biosci Biotechnol Biochem: 1999, 63(9);1528-34
[PubMed:10540738]
[WorldCat.org]
[DOI]
(P p)