Difference between revisions of "EtfB"
| Line 38: | Line 38: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | {{SubtiWiki regulon|[[FadR regulon]]}} | + | {{SubtiWiki regulon|[[CcpA regulon]]}}, {{SubtiWiki regulon|[[FadR regulon]]}} |
=The gene= | =The gene= | ||
| Line 107: | Line 107: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
** induced by long-chain fatty acids ([[FadR]]) {{PubMed|17189250}} | ** induced by long-chain fatty acids ([[FadR]]) {{PubMed|17189250}} | ||
| + | ** subject to carbon catabolite repression ([[CcpA]]-[[PtsH|HPr(Ser-P)]] {{PubMed|21398533}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
** [[FadR]]: transcription repression {{PubMed|17189250}} | ** [[FadR]]: transcription repression {{PubMed|17189250}} | ||
| + | ** [[CcpA]]-[[PtsH|HPr(Ser-P)]]: transcription repression {{PubMed|21398533}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 132: | Line 134: | ||
=References= | =References= | ||
| − | + | '''Additional publications:''' {{PubMed|21398533}} | |
<pubmed>17189250, 17919287, 17085570</pubmed> | <pubmed>17189250, 17919287, 17085570</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:57, 19 March 2011
- Description: electron transfer flavoprotein (beta subunit)
| Gene name | etfB |
| Synonyms | |
| Essential | no |
| Product | electron transfer flavoprotein (beta subunit) |
| Function | fatty acid degradation |
| Metabolic function and regulation of this protein in SubtiPathways: Fatty acid degradation | |
| MW, pI | 28 kDa, 4.187 |
| Gene length, protein length | 771 bp, 257 aa |
| Immediate neighbours | etfA, fadB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 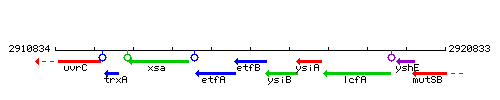
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
electron transport/ other, utilization of lipids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28530
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ETF beta-subunit/fixA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P94550
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- induced by long-chain fatty acids (FadR) PubMed
- subject to carbon catabolite repression (CcpA-HPr(Ser-P) PubMed
- Regulatory mechanism:
- FadR: transcription repression PubMed
- CcpA-HPr(Ser-P): transcription repression PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Yasutaro Fujita, Hiroshi Matsuoka, Kazutake Hirooka
Regulation of fatty acid metabolism in bacteria.
Mol Microbiol: 2007, 66(4);829-39
[PubMed:17919287]
[WorldCat.org]
[DOI]
(P p)
Hiroshi Matsuoka, Kazutake Hirooka, Yasutaro Fujita
Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation.
J Biol Chem: 2007, 282(8);5180-94
[PubMed:17189250]
[WorldCat.org]
[DOI]
(P p)
Chiara Barabesi, Alessandro Galizzi, Giorgio Mastromei, Mila Rossi, Elena Tamburini, Brunella Perito
Bacillus subtilis gene cluster involved in calcium carbonate biomineralization.
J Bacteriol: 2007, 189(1);228-35
[PubMed:17085570]
[WorldCat.org]
[DOI]
(P p)