Difference between revisions of "Eno"
(→References) |
(→Phenotypes of a mutant) |
||
| Line 39: | Line 39: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | |||
| + | essential [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | ||
=== Database entries === | === Database entries === | ||
Revision as of 12:04, 14 January 2009
- Description: enolase, glycolytic/ gluconeogenic enzyme
| Gene name | eno |
| Synonyms | |
| Essential | yes |
| Product | Enolase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| MW, pI | 46,4 kDa, 4.49 |
| Gene length, protein length | 1290 bp, 430 amino acids |
| Immediate neighbours | pgm, yvgK |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 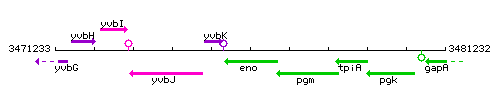
| |
Contents
The gene
Basic information
- Coordinates: 3475589 - 3476878
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O
- Protein family: enolase family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- substrate binding domain (366–369)
- Modification: phosphorylated during sporulation
- Cofactor(s): magnesium ion
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm.PubMed secreted. cell surface
Database entries
- Structure:
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Sigma factor: SigA
- Regulatory mechanism: repressor CggR PubMed, covalent substrate binding (Lys-339) causes inactivation, and is a possible signal for export of the protein
- Additional information:
Biological materials
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Commichau, F. M., Rothe, F. M., Herzberg, C., Wagner, E., Hellwig, D., Lehnik-Habrink, M., Hammer, E., Völker, U. & Stülke, J. Novel activities of glycolytic enzymes in Bacillus subtilis: Interactions with essential proteins involved in mRNA processing. subm.
- Ludwig, H., Homuth, G., Schmalisch, M., Dyka, F. M., Hecker, M., and Stülke, J. (2001) Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol Microbiol 41, 409-422.PubMed
- Jannière, L., Canceill, D., Suski, C., Kanga, S., Dalmais, B., Lestini, R., Monnier, A. F., Chapuis, J., Bolotin, A., Titok, M., Le Chatelier, E., and Ehrlich, S. D. (2007) Genetic evidence for a link between glycolysis and DNA replication. PLoS ONE 2, e447. PubMed
- Leyva-Vazquez, M. A., and Setlow, P. (1994) Cloning and nucleotide sequences of the genes encoding triose phosphate isomerase, phosphoglycerate mutase, and enolase from Bacillus subtilis. J Bacteriol 176: 3903-3910. PubMed