Difference between revisions of "Eno"
(→Expression and regulation) |
|||
| Line 20: | Line 20: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[pgm]]'', ''[[yvgK]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[pgm]]'', ''[[yvgK]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/eno_nucleotide.txt Gene sequence (+200bp) ]''' |
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/eno_protein.txt Protein sequence]''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:eno_context.gif]] |
|- | |- | ||
|} | |} | ||
Revision as of 15:38, 12 January 2009
- Description: Catalyzes the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate
| Gene name | eno |
| Synonyms | |
| Essential | yes |
| Product | Enolase |
| Function | lyase |
| MW, pI | 46,4 kDa, 4.49 |
| Gene length, protein length | 1290 bp, 430 amino acids |
| Immediate neighbours | pgm, yvgK |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 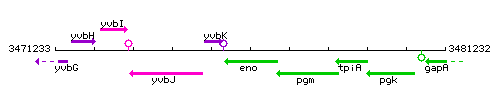
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O
- Protein family: enolase family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- substrate binding domain (366–369)
- Modification: phosphorylated during sporulation
- Cofactor(s): magnesium ion
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm.PubMed secreted. cell surface
Database entries
- Structure:
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Sigma factor: SigA
- Regulatory mechanism: repressor CggR PubMed, covalent substrate binding (Lys-339) causes inactivation, and is a possible signal for export of the protein
- Additional information:
Biological materials
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Commichau, F. M., Rothe, F. M., Herzberg, C., Wagner, E., Hellwig, D., Lehnik-Habrink, M., Hammer, E., Völker, U. & Stülke, J. Novel activities of glycolytic enzymes in Bacillus subtilis: Interactions with essential proteins involved in mRNA processing. subm.
- Ludwig, H., Homuth, G., Schmalisch, M., Dyka, F. M., Hecker, M., and Stülke, J. (2001) Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol Microbiol 41, 409-422.PubMed