Difference between revisions of "EfeO"
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | * '''Description:''' elemental iron uptake system ( | + | * '''Description:''' lipoprotein, elemental iron uptake system (binding protein), high affinity uptake of ferric iron (Fe(III)) <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || elemental iron uptake system ( | + | |style="background:#ABCDEF;" align="center"| '''Product''' || elemental iron uptake system (binding protein) |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || iron uptake | + | |style="background:#ABCDEF;" align="center"|'''Function''' || elemental iron uptake |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU38270 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU38270 efeO] |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/EfeO EfeO] |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=EfeO EfeO]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 42 kDa, 4.712 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 42 kDa, 4.712 | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1155 bp, 385 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1155 bp, 385 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[efeB]]'', ''[[efeU]]'' |
|- | |- | ||
|style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU38270 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU38270 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU38270 DNA_with_flanks] | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU38270 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU38270 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU38270 DNA_with_flanks] | ||
| Line 31: | Line 31: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=efeM_3927951_3929108_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:ywbM_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=efeM_3927951_3929108_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:ywbM_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU38270]] |
|- | |- | ||
|} | |} | ||
| Line 74: | Line 74: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** binds ferric iron (Fe(III)) with strong selectivity {{PubMed|23764491}} | ||
* '''Protein family:''' UPF0409 family (according to Swiss-Prot) | * '''Protein family:''' UPF0409 family (according to Swiss-Prot) | ||
| Line 92: | Line 93: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | ** | + | ** [[EfeO]]-[[EfeU]]-[[EfeB]] {{PubMed|23180473,23764491}} |
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
** membrane [http://www.ncbi.nlm.nih.gov/pubmed/18763711 PubMed] | ** membrane [http://www.ncbi.nlm.nih.gov/pubmed/18763711 PubMed] | ||
| − | + | ** extracytosolic lipoprotein {{PubMed|23764491}} | |
=== Database entries === | === Database entries === | ||
| Line 111: | Line 112: | ||
=Expression and regulation= | =Expression and regulation= | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | ** ''[[ | + | ** ''[[efeU]]-[[efeO]]-[[efeB]]'' {{PubMed|9353933}} |
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=efeM_3927951_3929108_-1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=efeM_3927951_3929108_-1 efeO] {{PubMed|22383849}} |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|23764491}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 124: | Line 125: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 1047 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 2113 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
| Line 145: | Line 148: | ||
=References= | =References= | ||
| − | <pubmed>16672620,9353933,12354229,18763711, 23180473</pubmed> | + | <pubmed>16672620,9353933,12354229,18763711, 23180473 23764491</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Latest revision as of 11:24, 17 April 2014
- Description: lipoprotein, elemental iron uptake system (binding protein), high affinity uptake of ferric iron (Fe(III))
| Gene name | ywbM |
| Synonyms | ipa-28d |
| Essential | no |
| Product | elemental iron uptake system (binding protein) |
| Function | elemental iron uptake |
| Gene expression levels in SubtiExpress: efeO | |
| Interactions involving this protein in SubtInteract: EfeO | |
| Metabolic function and regulation of this protein in SubtiPathways: EfeO | |
| MW, pI | 42 kDa, 4.712 |
| Gene length, protein length | 1155 bp, 385 aa |
| Immediate neighbours | efeB, efeU |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed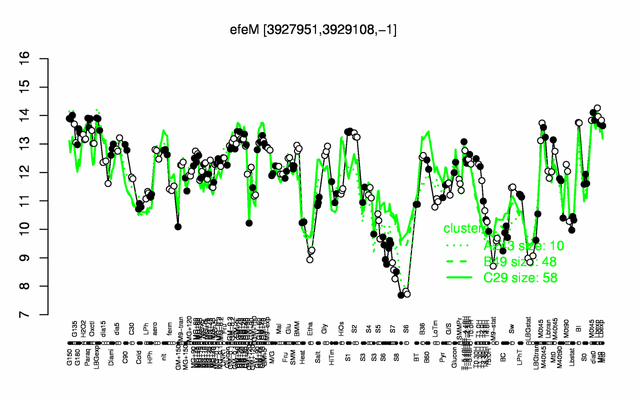
| |
Contents
Categories containing this gene/protein
transporters/ other, acquisition of iron, iron metabolism, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38270
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- binds ferric iron (Fe(III)) with strong selectivity PubMed
- Protein family: UPF0409 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39596
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Marcus Miethke, Carmine G Monteferrante, Mohamed A Marahiel, Jan Maarten van Dijl
The Bacillus subtilis EfeUOB transporter is essential for high-affinity acquisition of ferrous and ferric iron.
Biochim Biophys Acta: 2013, 1833(10);2267-78
[PubMed:23764491]
[WorldCat.org]
[DOI]
(P p)
Carmine G Monteferrante, Calum MacKichan, Elodie Marchadier, Maria-Victoria Prejean, Rut Carballido-López, Jan Maarten van Dijl
Mapping the twin-arginine protein translocation network of Bacillus subtilis.
Proteomics: 2013, 13(5);800-11
[PubMed:23180473]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Juliane Ollinger, Kyung-Bok Song, Haike Antelmann, Michael Hecker, John D Helmann
Role of the Fur regulon in iron transport in Bacillus subtilis.
J Bacteriol: 2006, 188(10);3664-73
[PubMed:16672620]
[WorldCat.org]
[DOI]
(P p)
Noel Baichoo, Tao Wang, Rick Ye, John D Helmann
Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon.
Mol Microbiol: 2002, 45(6);1613-29
[PubMed:12354229]
[WorldCat.org]
[DOI]
(P p)
E Presecan, I Moszer, L Boursier, H Cruz Ramos, V de la Fuente, M-F Hullo, C Lelong, S Schleich, A Sekowska, B H Song, G Villani, F Kunst, A Danchin, P Glaser
The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees).
Microbiology (Reading): 1997, 143 ( Pt 10);3313-3328
[PubMed:9353933]
[WorldCat.org]
[DOI]
(P p)