Difference between revisions of "EfeB"
(→Original publications) |
|||
| Line 162: | Line 162: | ||
<pubmed> 24140208 </pubmed> | <pubmed> 24140208 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>16672620,19180538,12354229, 19383693 9353933 9683469 15554971 21479178 18179421 22923395 23180473 23560556 23764491 23820555 </pubmed> | + | <pubmed>16672620,19180538,12354229, 19383693 9353933 9683469 15554971 21479178 18179421 22923395 23180473 23560556 23764491 23820555 24620988 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:38, 14 March 2014
- Description: elemental iron uptake system, heme peroxidase, converts ferrous iron (Fe(II) to ferric iron (FeIII)) for uptake by EfeO-EfeU, peroxide detoxification under microaerobic conditions
| Gene name | efeB |
| Synonyms | ipa-29d, ywbN |
| Essential | no |
| Product | heme peroxidase in elemental iron uptake |
| Function | ferrous iron conversion |
| Gene expression levels in SubtiExpress: efeB | |
| Interactions involving this protein in SubtInteract: EfeB | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 45 kDa, 8.64 |
| Gene length, protein length | 1248 bp, 416 aa |
| Immediate neighbours | ywbO, efeO |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 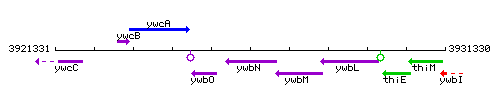
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed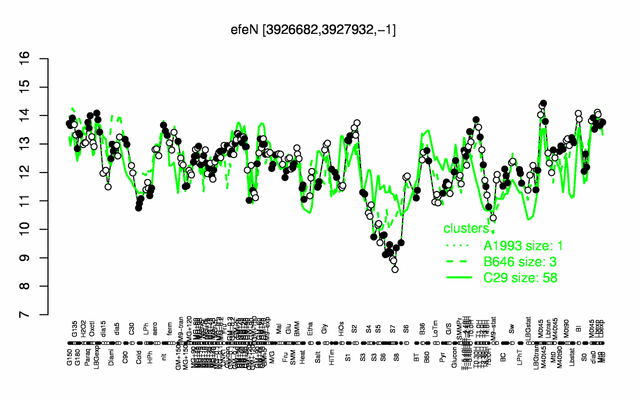
| |
Contents
Categories containing this gene/protein
acquisition of iron, iron metabolism, cell envelope stress proteins (controlled by SigM, V, W, X, Y), resistance against oxidative and electrophile stress, membrane proteins
This gene is a member of the following regulons
Fur regulon, SigM regulon, SigW regulon, SigX regulon
The gene
Basic information
- Locus tag: BSU38260
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: DyP-type peroxidase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39597
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Jan Maarten van Dijl, Groningen, Netherlands
Your additional remarks
References
Reviews
Vivianne J Goosens, Carmine G Monteferrante, Jan Maarten van Dijl
The Tat system of Gram-positive bacteria.
Biochim Biophys Acta: 2014, 1843(8);1698-706
[PubMed:24140208]
[WorldCat.org]
[DOI]
(P p)
Original publications
Ruihua Liu, Zhenqiang Zuo, Yingming Xu, Cunjiang Song, Hong Jiang, Chuanling Qiao, Ping Xu, Qixing Zhou, Chao Yang
Twin-arginine signal peptide of Bacillus subtilis YwbN can direct Tat-dependent secretion of methyl parathion hydrolase.
J Agric Food Chem: 2014, 62(13);2913-8
[PubMed:24620988]
[WorldCat.org]
[DOI]
(I p)
Ana Santos, Sónia Mendes, Vânia Brissos, Lígia O Martins
New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications.
Appl Microbiol Biotechnol: 2014, 98(5);2053-65
[PubMed:23820555]
[WorldCat.org]
[DOI]
(I p)
Marcus Miethke, Carmine G Monteferrante, Mohamed A Marahiel, Jan Maarten van Dijl
The Bacillus subtilis EfeUOB transporter is essential for high-affinity acquisition of ferrous and ferric iron.
Biochim Biophys Acta: 2013, 1833(10);2267-78
[PubMed:23764491]
[WorldCat.org]
[DOI]
(P p)
Murat Sezer, Ana Santos, Patrycja Kielb, Tiago Pinto, Ligia O Martins, Smilja Todorovic
Distinct structural and redox properties of the heme active site in bacterial dye decolorizing peroxidase-type peroxidases from two subfamilies: resonance Raman and electrochemical study.
Biochemistry: 2013, 52(18);3074-84
[PubMed:23560556]
[WorldCat.org]
[DOI]
(I p)
Carmine G Monteferrante, Calum MacKichan, Elodie Marchadier, Maria-Victoria Prejean, Rut Carballido-López, Jan Maarten van Dijl
Mapping the twin-arginine protein translocation network of Bacillus subtilis.
Proteomics: 2013, 13(5);800-11
[PubMed:23180473]
[WorldCat.org]
[DOI]
(I p)
Laxmi Krishnappa, Carmine G Monteferrante, Jan Maarten van Dijl
Degradation of the twin-arginine translocation substrate YwbN by extracytoplasmic proteases of Bacillus subtilis.
Appl Environ Microbiol: 2012, 78(21);7801-4
[PubMed:22923395]
[WorldCat.org]
[DOI]
(I p)
René van der Ploeg, Ulrike Mäder, Georg Homuth, Marc Schaffer, Emma L Denham, Carmine G Monteferrante, Marcus Miethke, Mohamed A Marahiel, Colin R Harwood, Theresa Winter, Michael Hecker, Haike Antelmann, Jan Maarten van Dijl
Environmental salinity determines the specificity and need for Tat-dependent secretion of the YwbN protein in Bacillus subtilis.
PLoS One: 2011, 6(3);e18140
[PubMed:21479178]
[WorldCat.org]
[DOI]
(I e)
Robyn T Eijlander, Magdalena A Kolbusz, Erwin M Berendsen, Oscar P Kuipers
Effects of altered TatC proteins on protein secretion efficiency via the twin-arginine translocation pathway of Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 6);1776-1785
[PubMed:19383693]
[WorldCat.org]
[DOI]
(P p)
Thijs R H M Kouwen, René van der Ploeg, Haike Antelmann, Michael Hecker, Georg Homuth, Ulrike Mäder, Jan Maarten van Dijl
Overflow of a hyper-produced secretory protein from the Bacillus Sec pathway into the Tat pathway for protein secretion as revealed by proteogenomics.
Proteomics: 2009, 9(4);1018-32
[PubMed:19180538]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Juliane Ollinger, Kyung-Bok Song, Haike Antelmann, Michael Hecker, John D Helmann
Role of the Fur regulon in iron transport in Bacillus subtilis.
J Bacteriol: 2006, 188(10);3664-73
[PubMed:16672620]
[WorldCat.org]
[DOI]
(P p)
Jan D H Jongbloed, Ulrike Grieger, Haike Antelmann, Michael Hecker, Reindert Nijland, Sierd Bron, Jan Maarten van Dijl
Two minimal Tat translocases in Bacillus.
Mol Microbiol: 2004, 54(5);1319-25
[PubMed:15554971]
[WorldCat.org]
[DOI]
(P p)
Noel Baichoo, Tao Wang, Rick Ye, John D Helmann
Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon.
Mol Microbiol: 2002, 45(6);1613-29
[PubMed:12354229]
[WorldCat.org]
[DOI]
(P p)
X Huang, K L Fredrick, J D Helmann
Promoter recognition by Bacillus subtilis sigmaW: autoregulation and partial overlap with the sigmaX regulon.
J Bacteriol: 1998, 180(15);3765-70
[PubMed:9683469]
[WorldCat.org]
[DOI]
(P p)
E Presecan, I Moszer, L Boursier, H Cruz Ramos, V de la Fuente, M-F Hullo, C Lelong, S Schleich, A Sekowska, B H Song, G Villani, F Kunst, A Danchin, P Glaser
The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees).
Microbiology (Reading): 1997, 143 ( Pt 10);3313-3328
[PubMed:9353933]
[WorldCat.org]
[DOI]
(P p)