Difference between revisions of "DtpT"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' proton-dependent di- and tripeptide transporter <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 97: | Line 97: | ||
* '''Structure:''' | * '''Structure:''' | ||
| + | ** [http://www.pdb.org/pdb/explore/explore.do?structureId=4ikv 4IKV], the protein from ''Geobacillus kaustophilus'' (65% identity, 87% similarity), {{PubMed|23798427}} | ||
* '''UniProt:''' [http://www.uniprot.org/uniprot/P94408 P94408] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P94408 P94408] | ||
| Line 142: | Line 143: | ||
=References= | =References= | ||
| − | <pubmed>11717292, 8458848 </pubmed> | + | <pubmed>11717292, 8458848 23798427 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:58, 26 November 2014
- Description: proton-dependent di- and tripeptide transporter
| Gene name | yclF |
| Synonyms | dtpT |
| Essential | no |
| Product | peptide transporter |
| Function | uptake of di- and tripeptides |
| Gene expression levels in SubtiExpress: yclF | |
| MW, pI | 53 kDa, 9.92 |
| Gene length, protein length | 1476 bp, 492 aa |
| Immediate neighbours | yclE, yclG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed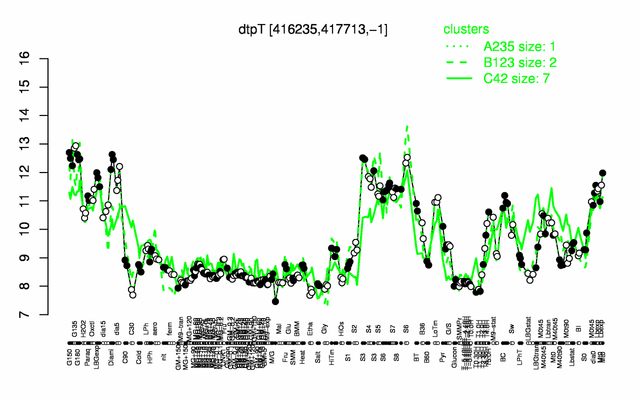
| |
Contents
Categories containing this gene/protein
transporters/ other, utilization of nitrogen sources other than amino acids, membrane proteins, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU03670
Phenotypes of a mutant
Database entries
- BsubCyc: BSU03670
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: POT family of secondary (PMF-dependent) transporters
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- BsubCyc: BSU03670
- UniProt: P94408
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Shintaro Doki, Hideaki E Kato, Nicolae Solcan, Masayo Iwaki, Michio Koyama, Motoyuki Hattori, Norihiko Iwase, Tomoya Tsukazaki, Yuji Sugita, Hideki Kandori, Simon Newstead, Ryuichiro Ishitani, Osamu Nureki
Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc Natl Acad Sci U S A: 2013, 110(28);11343-8
[PubMed:23798427]
[WorldCat.org]
[DOI]
(I p)
R Caldwell, R Sapolsky, W Weyler, R R Maile, S C Causey, E Ferrari
Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis.
J Bacteriol: 2001, 183(24);7329-40
[PubMed:11717292]
[WorldCat.org]
[DOI]
(P p)
E R Kunji, E J Smid, R Plapp, B Poolman, W N Konings
Di-tripeptides and oligopeptides are taken up via distinct transport mechanisms in Lactococcus lactis.
J Bacteriol: 1993, 175(7);2052-9
[PubMed:8458848]
[WorldCat.org]
[DOI]
(P p)