Difference between revisions of "DinB"
| Line 55: | Line 55: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU05630&redirect=T BSU05630] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/dinB.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/dinB.html] | ||
| Line 92: | Line 93: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU05630&redirect=T BSU05630] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 13:05, 2 April 2014
- Description: nuclease inhibitor
| Gene name | dinB |
| Synonyms | |
| Essential | no |
| Product | nuclease inhibitor |
| Function | response to DNA damage |
| Gene expression levels in SubtiExpress: dinB | |
| MW, pI | 19 kDa, 5.614 |
| Gene length, protein length | 516 bp, 172 aa |
| Immediate neighbours | ydgF, ydgG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 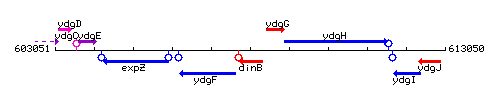
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed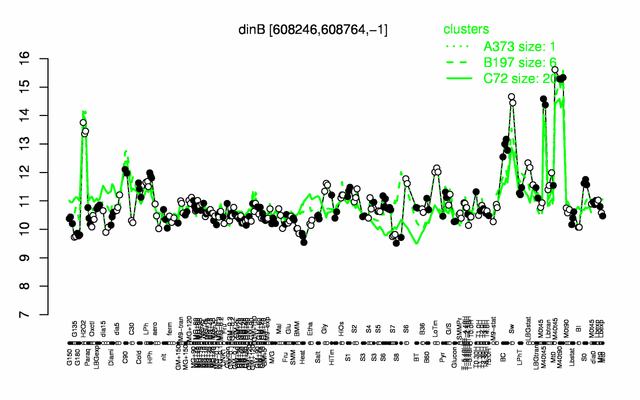
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU05630
Phenotypes of a mutant
Database entries
- BsubCyc: BSU05630
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: dinB family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU05630
- Structure:
- UniProt: Q02886
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Nora Au, Elke Kuester-Schoeck, Veena Mandava, Laura E Bothwell, Susan P Canny, Karen Chachu, Sierra A Colavito, Shakierah N Fuller, Eli S Groban, Laura A Hensley, Theresa C O'Brien, Amish Shah, Jessica T Tierney, Louise L Tomm, Thomas M O'Gara, Alexi I Goranov, Alan D Grossman, Charles M Lovett
Genetic composition of the Bacillus subtilis SOS system.
J Bacteriol: 2005, 187(22);7655-66
[PubMed:16267290]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
K W Winterling, D Chafin, J J Hayes, J Sun, A S Levine, R E Yasbin, R Woodgate
The Bacillus subtilis DinR binding site: redefinition of the consensus sequence.
J Bacteriol: 1998, 180(8);2201-11
[PubMed:9555905]
[WorldCat.org]
[DOI]
(P p)
M C Miller, J B Resnick, B T Smith, C M Lovett
The bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. Purification and characterization of the DinR protein.
J Biol Chem: 1996, 271(52);33502-8
[PubMed:8969214]
[WorldCat.org]
(P p)
C M Lovett, K C Cho, T M O'Gara
Purification of an SOS repressor from Bacillus subtilis.
J Bacteriol: 1993, 175(21);6842-9
[PubMed:8226626]
[WorldCat.org]
[DOI]
(P p)
D L Cheo, K W Bayles, R E Yasbin
Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis.
J Bacteriol: 1991, 173(5);1696-703
[PubMed:1847907]
[WorldCat.org]
[DOI]
(P p)