Difference between revisions of "Des"
(→References) |
|||
| Line 16: | Line 16: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU19180 des] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU19180 des] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=Des Des] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_deg.html Fatty acid degradation]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_deg.html Fatty acid degradation]''' | ||
Revision as of 08:35, 12 November 2013
- Description: phospholipid desaturase
| Gene name | des |
| Synonyms | yocE |
| Essential | no |
| Product | phospholipid desaturase |
| Function | adaptation of membrane fluidity at low temperatures |
| Gene expression levels in SubtiExpress: des | |
| Interactions involving this protein in SubtInteract: Des | |
| Metabolic function and regulation of this protein in SubtiPathways: Fatty acid degradation | |
| MW, pI | 40 kDa, 9.88 |
| Gene length, protein length | 1056 bp, 352 aa |
| Immediate neighbours | yocD, desK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 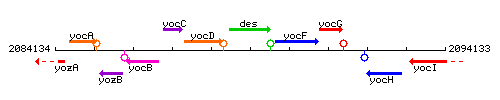
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed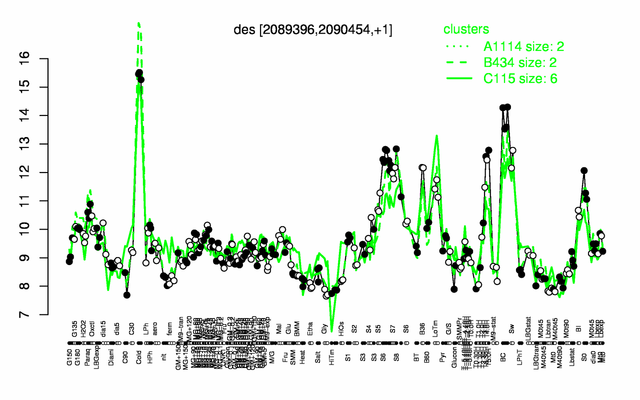
| |
Contents
Categories containing this gene/protein
lipid metabolism/ other, cold stress proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU19180
Phenotypes of a mutant
- cold-sensitive PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- in strains evolved for growth at low pressure, des expression is significantly increased as compared to the wild type, this allows growth at low pressure PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: introduces double bonds into long chain fatty acids PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: integral membrane protein PubMed
Database entries
- Structure:
- UniProt: O34653
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: des PubMed
- Additional information:
- in strains evolved for growth at low pressure, des expression is significantly increased as compared to the wild type PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Diego de Mendoza, Universidad Nacional de Rosario, Argentine homepage
Your additional remarks
References
Reviews
John Shanklin, Jodie E Guy, Girish Mishra, Ylva Lindqvist
Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes.
J Biol Chem: 2009, 284(28);18559-63
[PubMed:19363032]
[WorldCat.org]
[DOI]
(P p)
Original publications
Patricia Fajardo-Cavazos, Samantha M Waters, Andrew C Schuerger, Sheeja George, James J Marois, Wayne L Nicholson
Evolution of Bacillus subtilis to enhanced growth at low pressure: up-regulated transcription of des-desKR, encoding the fatty acid desaturase system.
Astrobiology: 2012, 12(3);258-70
[PubMed:22416764]
[WorldCat.org]
[DOI]
(I p)
Lorena Chazarreta-Cifre, Leticia Martiarena, Diego de Mendoza, Silvia G Altabe
Role of ferredoxin and flavodoxins in Bacillus subtilis fatty acid desaturation.
J Bacteriol: 2011, 193(16);4043-8
[PubMed:21665975]
[WorldCat.org]
[DOI]
(I p)
Jana Beranová, María C Mansilla, Diego de Mendoza, Dana Elhottová, Ivo Konopásek
Differences in cold adaptation of Bacillus subtilis under anaerobic and aerobic conditions.
J Bacteriol: 2010, 192(16);4164-71
[PubMed:20581210]
[WorldCat.org]
[DOI]
(I p)
Natalia Martin, Esteban Lombardía, Silvia G Altabe, Diego de Mendoza, María C Mansilla
A lipA (yutB) mutant, encoding lipoic acid synthase, provides insight into the interplay between branched-chain and unsaturated fatty acid biosynthesis in Bacillus subtilis.
J Bacteriol: 2009, 191(24);7447-55
[PubMed:19820084]
[WorldCat.org]
[DOI]
(I p)
Anna-Barbara Hachmann, Esther R Angert, John D Helmann
Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin.
Antimicrob Agents Chemother: 2009, 53(4);1598-609
[PubMed:19164152]
[WorldCat.org]
[DOI]
(I p)
Jana Beranová, Małgorzata Jemioła-Rzemińska, Dana Elhottová, Kazimierz Strzałka, Ivo Konopásek
Metabolic control of the membrane fluidity in Bacillus subtilis during cold adaptation.
Biochim Biophys Acta: 2008, 1778(2);445-53
[PubMed:18154726]
[WorldCat.org]
[DOI]
(P p)
Silvia G Altabe, Pablo Aguilar, Gerardo M Caballero, Diego de Mendoza
The Bacillus subtilis acyl lipid desaturase is a delta5 desaturase.
J Bacteriol: 2003, 185(10);3228-31
[PubMed:12730185]
[WorldCat.org]
[DOI]
(P p)
Carsten L Beckering, Leif Steil, Michael H W Weber, Uwe Völker, Mohamed A Marahiel
Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis.
J Bacteriol: 2002, 184(22);6395-402
[PubMed:12399512]
[WorldCat.org]
[DOI]
(P p)
Larisa E Cybulski, Daniela Albanesi, María C Mansilla, Silvia Altabe, Pablo S Aguilar, Diego de Mendoza
Mechanism of membrane fluidity optimization: isothermal control of the Bacillus subtilis acyl-lipid desaturase.
Mol Microbiol: 2002, 45(5);1379-88
[PubMed:12207704]
[WorldCat.org]
[DOI]
(P p)
P S Aguilar, A M Hernandez-Arriaga, L E Cybulski, A C Erazo, D de Mendoza
Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis.
EMBO J: 2001, 20(7);1681-91
[PubMed:11285232]
[WorldCat.org]
[DOI]
(P p)
M H Weber, W Klein, L Müller, U M Niess, M A Marahiel
Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock.
Mol Microbiol: 2001, 39(5);1321-9
[PubMed:11251847]
[WorldCat.org]
[DOI]
(P p)
P S Aguilar, P Lopez, D de Mendoza
Transcriptional control of the low-temperature-inducible des gene, encoding the delta5 desaturase of Bacillus subtilis.
J Bacteriol: 1999, 181(22);7028-33
[PubMed:10559169]
[WorldCat.org]
[DOI]
(P p)
P S Aguilar, J E Cronan, D de Mendoza
A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase.
J Bacteriol: 1998, 180(8);2194-200
[PubMed:9555904]
[WorldCat.org]
[DOI]
(P p)