Difference between revisions of "Dat"
| Line 25: | Line 25: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yheN]]'', ''[[nhaC]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yheN]]'', ''[[nhaC]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU09670 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU09670 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU09670 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU09670 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU09670 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU09670 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:dat_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:dat_context.gif]] | ||
Revision as of 09:48, 14 May 2013
(Another gene coding for O6-methylguanine DNA alkyltransferase was formely known as dat, it is now called ogt)
| Gene name | dat |
| Synonyms | yheM |
| Essential | no |
| Product | D-alanine aminotransferase |
| Function | peptidoglycan precursor biosynthesis |
| Gene expression levels in SubtiExpress: dat | |
| Metabolic function and regulation of this protein in SubtiPathways: Cell wall | |
| MW, pI | 31 kDa, 4.871 |
| Gene length, protein length | 846 bp, 282 aa |
| Immediate neighbours | yheN, nhaC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 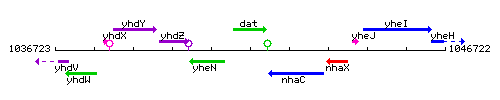
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed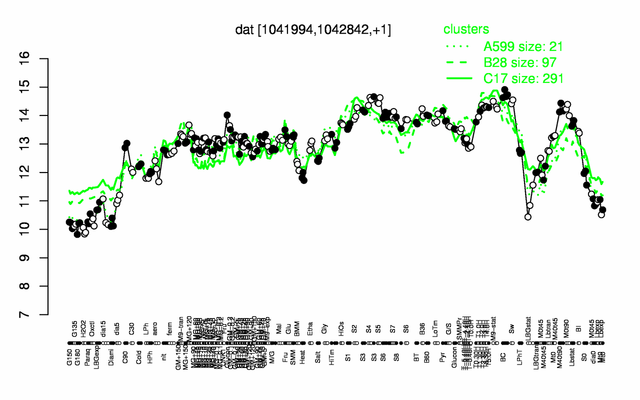
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU09670
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-alanine + 2-oxoglutarate = pyruvate + D-glutamate (according to Swiss-Prot)
- Protein family: class-IV pyridoxal-phosphate-dependent aminotransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1G2W (E177S Mutant, Geobacillus stearothermophilus, 43% identity)
- UniProt: O07597
- KEGG entry: [2]
- E.C. number: 2.6.1.21
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Le Thi Tam, Christine Eymann, Haike Antelmann, Dirk Albrecht, Michael Hecker
Global gene expression profiling of Bacillus subtilis in response to ammonium and tryptophan starvation as revealed by transcriptome and proteome analysis.
J Mol Microbiol Biotechnol: 2007, 12(1-2);121-30
[PubMed:17183219]
[WorldCat.org]
[DOI]
(P p)
P P Taylor, I G Fotheringham
Nucleotide sequence of the Bacillus licheniformis ATCC 10716 dat gene and comparison of the predicted amino acid sequence with those of other bacterial species.
Biochim Biophys Acta: 1997, 1350(1);38-40
[PubMed:9003455]
[WorldCat.org]
[DOI]
(P p)