Difference between revisions of "DacC"
| Line 22: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ppsA]]'', ''[[galM]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ppsA]]'', ''[[galM]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU18350 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU18350 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU18350 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU18350 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU18350 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU18350 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:dacC_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:dacC_context.gif]] | ||
Revision as of 10:22, 14 May 2013
- Description: penicillin-binding protein 4A, D-alanyl-D-alanine carboxypeptidase
| Gene name | dacC |
| Synonyms | pbp |
| Essential | no |
| Product | penicillin-binding protein 4A, D-alanyl-D-alanine carboxypeptidase |
| Function | carboxypeptidase |
| Gene expression levels in SubtiExpress: dacC | |
| MW, pI | 52 kDa, 5.413 |
| Gene length, protein length | 1473 bp, 491 aa |
| Immediate neighbours | ppsA, galM |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 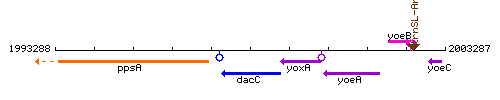
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed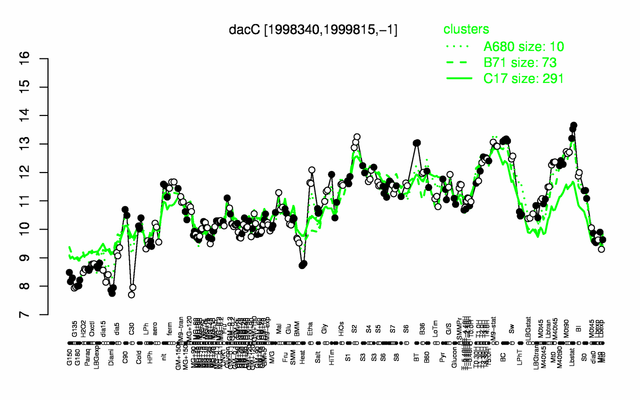
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU18350
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relexed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: cleavage ofD-Ala-D-Ala interpeptide bridges in peptidoglycan PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- may be part of the cell wall biosynthetic complex PubMed
Database entries
- UniProt: P39844
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Venkatesh V Nemmara, S A Adediran, Kinjal Dave, Colette Duez, R F Pratt
Dual substrate specificity of Bacillus subtilis PBP4a.
Biochemistry: 2013, 52(15);2627-37
[PubMed:23560856]
[WorldCat.org]
[DOI]
(I p)
Venkatesh V Nemmara, Liudmila Dzhekieva, Kumar Subarno Sarkar, S A Adediran, Colette Duez, Robert A Nicholas, R F Pratt
Substrate specificity of low-molecular mass bacterial DD-peptidases.
Biochemistry: 2011, 50(46);10091-101
[PubMed:22029692]
[WorldCat.org]
[DOI]
(I p)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Colette Duez, Astrid Zervosen, Nathalie Teller, Rémy Melkonian, Emmanuel Banzubazé, Fabrice Bouillenne, André Luxen, Jean-Marie Frère
Characterization of the proteins encoded by the Bacillus subtilis yoxA-dacC operon.
FEMS Microbiol Lett: 2009, 300(1);42-7
[PubMed:19758330]
[WorldCat.org]
[DOI]
(I p)
Eric Sauvage, Colette Duez, Raphaël Herman, Frédéric Kerff, Stephanie Petrella, John W Anderson, S A Adediran, R F Pratt, Jean-Marie Frère, Paulette Charlier
Crystal structure of the Bacillus subtilis penicillin-binding protein 4a, and its complex with a peptidoglycan mimetic peptide.
J Mol Biol: 2007, 371(2);528-39
[PubMed:17582436]
[WorldCat.org]
[DOI]
(P p)
Dirk-Jan Scheffers, Laura J F Jones, Jeffery Errington
Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis.
Mol Microbiol: 2004, 51(3);749-64
[PubMed:14731276]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, J Bernhardt, U Gerth, D Höper, T Koburger, U Völker, M Hecker
Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization.
J Bacteriol: 1999, 181(18);5718-24
[PubMed:10482513]
[WorldCat.org]
[DOI]
(P p)
D L Popham, M E Gilmore, P Setlow
Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties.
J Bacteriol: 1999, 181(1);126-32
[PubMed:9864321]
[WorldCat.org]
[DOI]
(P p)
L B Pedersen, T Murray, D L Popham, P Setlow
Characterization of dacC, which encodes a new low-molecular-weight penicillin-binding protein in Bacillus subtilis.
J Bacteriol: 1998, 180(18);4967-73
[PubMed:9733705]
[WorldCat.org]
[DOI]
(P p)