Difference between revisions of "DacB"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 111: | Line 107: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=dacB_2423383_2424531_-1 dacB] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=dacB_2423383_2424531_-1 dacB] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigE]] {{PubMed|15699190,7528199}} | + | * '''[[Sigma factor]]:''' [[SigE]] {{PubMed|15699190,7528199}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 141: | Line 137: | ||
=References= | =References= | ||
| − | + | <pubmed>10498740,9864321,1548223,7642500,14731276,19542328 ,7528199 15699190 10383963 22123250</pubmed> | |
| − | <pubmed>10498740,9864321,1548223,7642500,14731276,19542328 ,7528199 15699190 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:22, 10 May 2013
- Description: sporulation-specific penicillin-binding protein 5, D-alanyl-D-alanine carboxypeptidase
| Gene name | dacB |
| Synonyms | |
| Essential | no |
| Product | penicillin-binding protein 5, D-alanyl-D-alanine carboxypeptidase |
| Function | carboxypeptidase |
| Gene expression levels in SubtiExpress: dacB | |
| MW, pI | 42 kDa, 9.725 |
| Gene length, protein length | 1146 bp, 382 aa |
| Immediate neighbours | spmA, ypuI |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 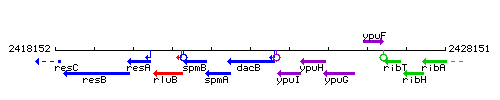
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed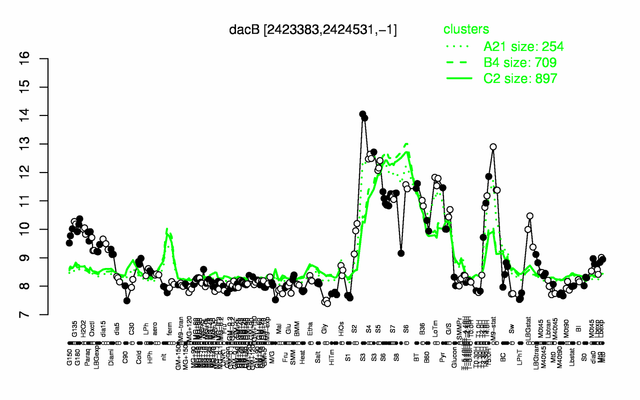
| |
Contents
Categories containing this gene/protein
cell wall synthesis, sporulation proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23190
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: modifies degree of cross-linking of glycan strands in peptidoglycan
- Protein family: peptidase S11 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure: 3MFD (beta-lactamase superfamily domain)
- UniProt: P35150
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Pengfei Zhang, Stacy Thomas, Yong-qing Li, Peter Setlow
Effects of cortex peptidoglycan structure and cortex hydrolysis on the kinetics of Ca(2+)-dipicolinic acid release during Bacillus subtilis spore germination.
J Bacteriol: 2012, 194(3);646-52
[PubMed:22123250]
[WorldCat.org]
[DOI]
(I p)
Ralf Moeller, Peter Setlow, Günther Reitz, Wayne L Nicholson
Roles of small, acid-soluble spore proteins and core water content in survival of Bacillus subtilis spores exposed to environmental solar UV radiation.
Appl Environ Microbiol: 2009, 75(16);5202-8
[PubMed:19542328]
[WorldCat.org]
[DOI]
(I p)
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
Dirk-Jan Scheffers, Laura J F Jones, Jeffery Errington
Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis.
Mol Microbiol: 2004, 51(3);749-64
[PubMed:14731276]
[WorldCat.org]
[DOI]
(P p)
D L Popham, J Meador-Parton, C E Costello, P Setlow
Spore peptidoglycan structure in a cwlD dacB double mutant of Bacillus subtilis.
J Bacteriol: 1999, 181(19);6205-9
[PubMed:10498740]
[WorldCat.org]
[DOI]
(P p)
A Atrih, G Bacher, G Allmaier, M P Williamson, S J Foster
Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation.
J Bacteriol: 1999, 181(13);3956-66
[PubMed:10383963]
[WorldCat.org]
[DOI]
(P p)
D L Popham, M E Gilmore, P Setlow
Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties.
J Bacteriol: 1999, 181(1);126-32
[PubMed:9864321]
[WorldCat.org]
[DOI]
(P p)
D L Popham, B Illades-Aguiar, P Setlow
The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration.
J Bacteriol: 1995, 177(16);4721-9
[PubMed:7642500]
[WorldCat.org]
[DOI]
(P p)
E B Simpson, T W Hancock, C E Buchanan
Transcriptional control of dacB, which encodes a major sporulation-specific penicillin-binding protein.
J Bacteriol: 1994, 176(24);7767-9
[PubMed:7528199]
[WorldCat.org]
[DOI]
(P p)
C E Buchanan, M L Ling
Isolation and sequence analysis of dacB, which encodes a sporulation-specific penicillin-binding protein in Bacillus subtilis.
J Bacteriol: 1992, 174(6);1717-25
[PubMed:1548223]
[WorldCat.org]
[DOI]
(P p)