Difference between revisions of "CwlS"
(→This gene is a member of the following regulons) |
(→This gene is a member of the following regulons) |
||
| Line 36: | Line 36: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | {{SubtiWiki regulon|[[ | + | {{SubtiWiki regulon|[[Abh regulon]]}}, |
| − | {{SubtiWiki regulon|[[AbrB regulon]]}} | + | {{SubtiWiki regulon|[[AbrB regulon]]}}, |
| + | {{SubtiWiki regulon|[[CcpA regulon]]}} | ||
=The gene= | =The gene= | ||
Revision as of 13:45, 14 July 2011
- Description: D,L-endopeptidase, peptidoglycan hydrolase
| Gene name | cwlS |
| Synonyms | yojL |
| Essential | no |
| Product | D,L-endopeptidase, peptidoglycan hydrolase |
| Function | cell wall metabolism |
| MW, pI | 44 kDa, 10.438 |
| Gene length, protein length | 1242 bp, 414 aa |
| Immediate neighbours | yojM, yojK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 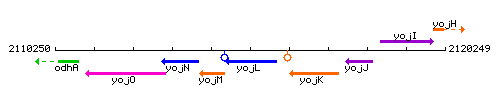
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, membrane proteins
This gene is a member of the following regulons
Abh regulon, AbrB regulon, CcpA regulon
The gene
Basic information
- Locus tag: BSU19410
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of gamma-D-glutamyl bonds to the L-terminus (position 7) of meso-diaminopimelic acid (meso-A2pm) in 7-(L-Ala-gamma-D-Glu)-meso-A2pm and 7-(L-Ala-gamma-D-Glu)-7-(D-Ala)-meso-A2pm (according to Swiss-Prot)
- Protein family: Cu-Zn superoxide dismutase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O31852
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Tatsuya Fukushima, Anahita Afkham, Shin-Ichirou Kurosawa, Taichi Tanabe, Hiroki Yamamoto, Junichi Sekiguchi
A new D,L-endopeptidase gene product, YojL (renamed CwlS), plays a role in cell separation with LytE and LytF in Bacillus subtilis.
J Bacteriol: 2006, 188(15);5541-50
[PubMed:16855244]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)