Difference between revisions of "CtaB"
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[ctaB]]-[[ctaC]]-[[ctaD]]-[[ctaE]]-[[ctaF]]-[[ctaG]]'' {{PubMed|9829923}} |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** expressed under anaerobic conditions ([[ResD]]) | + | ** expressed under anaerobic conditions ([[ResD]]) {{PubMed|9829923}} |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | ** [[ResD]]: transcription activation | + | ** [[ResD]]: transcription activation {{PubMed|9829923}} |
* '''Additional information:''' | * '''Additional information:''' | ||
Revision as of 20:18, 8 October 2009
- Description: heme O synthase (major enzyme)
| Gene name | ctaB |
| Synonyms | |
| Essential | no |
| Product | heme O synthase (major enzyme) |
| Function | heme biosynthesis |
| MW, pI | 33 kDa, 9.566 |
| Gene length, protein length | 915 bp, 305 aa |
| Immediate neighbours | ctaA, ctaC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 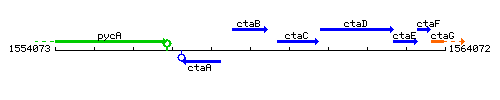
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU14880
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ubiA prenyltransferase family, Protoheme IX farnesyltransferase subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P24009
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Tatsushi Mogi
Over-expression and characterization of Bacillus subtilis heme O synthase.
J Biochem: 2009, 145(5);669-75
[PubMed:19204012]
[WorldCat.org]
[DOI]
(I p)
Brienne M Brown, Zhihong Wang, Kenneth R Brown, Julia A Cricco, Eric L Hegg
Heme O synthase and heme A synthase from Bacillus subtilis and Rhodobacter sphaeroides interact in Escherichia coli.
Biochemistry: 2004, 43(42);13541-8
[PubMed:15491161]
[WorldCat.org]
[DOI]
(P p)
X Liu, H W Taber
Catabolite regulation of the Bacillus subtilis ctaBCDEF gene cluster.
J Bacteriol: 1998, 180(23);6154-63
[PubMed:9829923]
[WorldCat.org]
[DOI]
(P p)
B Svensson, M Lübben, L Hederstedt
Bacillus subtilis CtaA and CtaB function in haem A biosynthesis.
Mol Microbiol: 1993, 10(1);193-201
[PubMed:7968515]
[WorldCat.org]
[DOI]
(P p)