Difference between revisions of "CspB"
(→References) |
|||
| Line 36: | Line 36: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[RNA chaperones]]}}, | {{SubtiWiki category|[[RNA chaperones]]}}, | ||
| − | {{SubtiWiki category|[[cold stress proteins]]}} | + | {{SubtiWiki category|[[cold stress proteins]]}}, |
| + | [[most abundant proteins]] | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 64: | Line 61: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 82: | Line 76: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 117: | Line 111: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cspB_984262_984465_-1 cspB] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cspB_984262_984465_-1 cspB] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|1400185}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|1400185}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 124: | Line 118: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation {{PubMed|17981983}} | + | * '''Additional information:''' |
| + | ** subject to Clp-dependent proteolysis upon glucose starvation {{PubMed|17981983}} | ||
| + | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
=Biological materials = | =Biological materials = | ||
| Line 147: | Line 143: | ||
=References= | =References= | ||
| − | <pubmed>12399512,12171653,11752341,11717297,12838604,8321288,8321289,9533624,9379903,8294017,8755892,9914312,11591689,7476164,1409560,16352840,9920884,1400185,12427936,8307174,7703860,16352840, 22128343 23199363 21124848</pubmed> | + | <pubmed>12399512,12171653,11752341,11717297,12838604,8321288,8321289,9533624,9379903,8294017,8755892,9914312, 11591689,7476164,1409560,16352840,9920884,1400185,12427936,8307174,7703860,16352840, 22128343 23199363 15378759 21124848</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:01, 5 March 2014
- Description: major cold-shock protein
| Gene name | cspB |
| Synonyms | |
| Essential | no |
| Product | major cold-shock protein |
| Function | RNA chaperone |
| Gene expression levels in SubtiExpress: cspB | |
| Interactions involving this protein in SubtInteract: CspB | |
| MW, pI | 7 kDa, 4.341 |
| Gene length, protein length | 201 bp, 67 aa |
| Immediate neighbours | yhcI, yhcJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 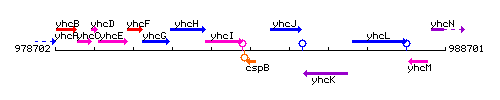
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed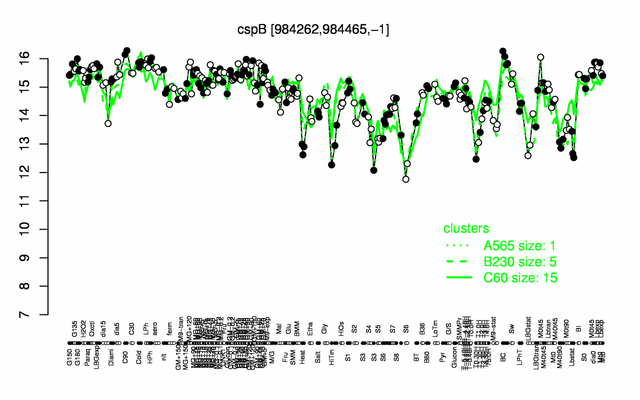
| |
Contents
Categories containing this gene/protein
RNA chaperones, cold stress proteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU09100
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot), cytoplasma, colocalizes with the ribosomes PubMed
Database entries
- UniProt: P32081
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon: cspB PubMed
- Regulation:
- induced upon cold shock PubMed
- Regulatory mechanism:
- Additional information:
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Mohamed Marahiel, Marburg University, Germany homepage
Your additional remarks
References
Letal I Salzberg, Leagh Powell, Karsten Hokamp, Eric Botella, David Noone, Kevin M Devine
The WalRK (YycFG) and σ(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells.
Mol Microbiol: 2013, 87(1);180-95
[PubMed:23199363]
[WorldCat.org]
[DOI]
(I p)
Rolf Sachs, Klaas E A Max, Udo Heinemann, Jochen Balbach
RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
RNA: 2012, 18(1);65-76
[PubMed:22128343]
[WorldCat.org]
[DOI]
(I p)
Anja Buttstedt, Reno Winter, Mirko Sackewitz, Gerd Hause, Franz-Xaver Schmid, Elisabeth Schwarz
Influence of the stability of a fused protein and its distance to the amyloidogenic segment on fibril formation.
PLoS One: 2010, 5(11);e15436
[PubMed:21124848]
[WorldCat.org]
[DOI]
(I e)
Karen Hunger, Carsten L Beckering, Frank Wiegeshoff, Peter L Graumann, Mohamed A Marahiel
Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis.
J Bacteriol: 2006, 188(1);240-8
[PubMed:16352840]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Michael H Weber, Mohamed A Marahiel
Bacterial cold shock responses.
Sci Prog: 2003, 86(Pt 1-2);9-75
[PubMed:12838604]
[WorldCat.org]
[DOI]
(P p)
Tanja Kaan, Georg Homuth, Ulrike Mäder, Julia Bandow, Thomas Schweder
Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response.
Microbiology (Reading): 2002, 148(Pt 11);3441-3455
[PubMed:12427936]
[WorldCat.org]
[DOI]
(P p)
Carsten L Beckering, Leif Steil, Michael H W Weber, Uwe Völker, Mohamed A Marahiel
Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis.
J Bacteriol: 2002, 184(22);6395-402
[PubMed:12399512]
[WorldCat.org]
[DOI]
(P p)
Michael H W Weber, Mohamed A Marahiel
Coping with the cold: the cold shock response in the Gram-positive soil bacterium Bacillus subtilis.
Philos Trans R Soc Lond B Biol Sci: 2002, 357(1423);895-907
[PubMed:12171653]
[WorldCat.org]
[DOI]
(P p)
Michael H W Weber, Ingo Fricke, Niclas Doll, Mohamed A Marahiel
CSDBase: an interactive database for cold shock domain-containing proteins and the bacterial cold shock response.
Nucleic Acids Res: 2002, 30(1);375-8
[PubMed:11752341]
[WorldCat.org]
[DOI]
(I p)
M H Weber, C L Beckering, M A Marahiel
Complementation of cold shock proteins by translation initiation factor IF1 in vivo.
J Bacteriol: 2001, 183(24);7381-6
[PubMed:11717297]
[WorldCat.org]
[DOI]
(P p)
M H Weber, A V Volkov, I Fricke, M A Marahiel, P L Graumann
Localization of cold shock proteins to cytosolic spaces surrounding nucleoids in Bacillus subtilis depends on active transcription.
J Bacteriol: 2001, 183(21);6435-43
[PubMed:11591689]
[WorldCat.org]
[DOI]
(P p)
T Schindler, P L Graumann, D Perl, S Ma, F X Schmid, M A Marahiel
The family of cold shock proteins of Bacillus subtilis. Stability and dynamics in vitro and in vivo.
J Biol Chem: 1999, 274(6);3407-13
[PubMed:9920884]
[WorldCat.org]
[DOI]
(P p)
P L Graumann, M A Marahiel
Cold shock proteins CspB and CspC are major stationary-phase-induced proteins in Bacillus subtilis.
Arch Microbiol: 1999, 171(2);135-8
[PubMed:9914312]
[WorldCat.org]
[DOI]
(P p)
T Schindler, D Perl, P Graumann, V Sieber, M A Marahiel, F X Schmid
Surface-exposed phenylalanines in the RNP1/RNP2 motif stabilize the cold-shock protein CspB from Bacillus subtilis.
Proteins: 1998, 30(4);401-6
[PubMed:9533624]
[WorldCat.org]
[DOI]
(P p)
P Graumann, T M Wendrich, M H Weber, K Schröder, M A Marahiel
A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures.
Mol Microbiol: 1997, 25(4);741-56
[PubMed:9379903]
[WorldCat.org]
[DOI]
(P p)
P Graumann, K Schröder, R Schmid, M A Marahiel
Cold shock stress-induced proteins in Bacillus subtilis.
J Bacteriol: 1996, 178(15);4611-9
[PubMed:8755892]
[WorldCat.org]
[DOI]
(P p)
K Schröder, P Graumann, A Schnuchel, T A Holak, M A Marahiel
Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif.
Mol Microbiol: 1995, 16(4);699-708
[PubMed:7476164]
[WorldCat.org]
[DOI]
(P p)
G I Makhatadze, M A Marahiel
Effect of pH and phosphate ions on self-association properties of the major cold-shock protein from Bacillus subtilis.
Protein Sci: 1994, 3(11);2144-7
[PubMed:7703860]
[WorldCat.org]
[DOI]
(P p)
P Graumann, M A Marahiel
The major cold shock protein of Bacillus subtilis CspB binds with high affinity to the ATTGG- and CCAAT sequences in single stranded oligonucleotides.
FEBS Lett: 1994, 338(2);157-60
[PubMed:8307174]
[WorldCat.org]
[DOI]
(P p)
K Schröder, P Zuber, G Willimsky, B Wagner, M A Marahiel
Mapping of the Bacillus subtilis cspB gene and cloning of its homologs in thermophilic, mesophilic and psychrotrophic bacilli.
Gene: 1993, 136(1-2);277-80
[PubMed:8294017]
[WorldCat.org]
[DOI]
(P p)
A Schnuchel, R Wiltscheck, M Czisch, M Herrler, G Willimsky, P Graumann, M A Marahiel, T A Holak
Structure in solution of the major cold-shock protein from Bacillus subtilis.
Nature: 1993, 364(6433);169-71
[PubMed:8321289]
[WorldCat.org]
[DOI]
(P p)
H Schindelin, M A Marahiel, U Heinemann
Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein.
Nature: 1993, 364(6433);164-8
[PubMed:8321288]
[WorldCat.org]
[DOI]
(P p)
G Willimsky, H Bang, G Fischer, M A Marahiel
Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures.
J Bacteriol: 1992, 174(20);6326-35
[PubMed:1400185]
[WorldCat.org]
[DOI]
(P p)
H Schindelin, M Herrler, G Willimsky, M A Marahiel, U Heinemann
Overproduction, crystallization, and preliminary X-ray diffraction studies of the major cold shock protein from Bacillus subtilis, CspB.
Proteins: 1992, 14(1);120-4
[PubMed:1409560]
[WorldCat.org]
[DOI]
(P p)