Difference between revisions of "ComFA"
(→Expression and regulation) |
|||
| Line 87: | Line 87: | ||
=Expression and regulation= | =Expression and regulation= | ||
| + | * '''Operon:''' ''[[comFA]]-[[comFB]]-[[comFC]]-[[yvyF]]-[[flgM]]-[[yvyG]]-[[flgK]]-[[flgL]]'' {{PubMed|8412657}} | ||
| − | * ''' | + | * '''[[Sigma factor]]:''' ''[[comFA]]'': [[SigA]] {{PubMed|8412657}} |
| − | |||
| − | |||
* '''Regulation:''' | * '''Regulation:''' | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[ComK]]: transcription activation {{PubMed|8412657}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 21:45, 30 December 2009
- Description: late competence protein required for DNA binding and uptake
| Gene name | comFA |
| Synonyms | |
| Essential | no |
| Product | DNA-binding protein |
| Function | genetic transformation |
| MW, pI | 52 kDa, 9.824 |
| Gene length, protein length | 1389 bp, 463 aa |
| Immediate neighbours | comFB, yviA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 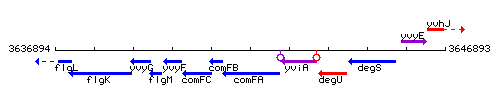
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU35470
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: helicase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
Database entries
- Structure:
- UniProt: P39145
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Naomi Kramer, Jeanette Hahn, David Dubnau
Multiple interactions among the competence proteins of Bacillus subtilis.
Mol Microbiol: 2007, 65(2);454-64
[PubMed:17630974]
[WorldCat.org]
[DOI]
(P p)
Jeanette Hahn, Berenike Maier, Bert Jan Haijema, Michael Sheetz, David Dubnau
Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis.
Cell: 2005, 122(1);59-71
[PubMed:16009133]
[WorldCat.org]
[DOI]
(P p)
Hanne Jarmer, Randy Berka, Steen Knudsen, Hans H Saxild
Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions.
FEMS Microbiol Lett: 2002, 206(2);197-200
[PubMed:11814663]
[WorldCat.org]
[DOI]
(P p)
J A Londoño-Vallejo, D Dubnau
Mutation of the putative nucleotide binding site of the Bacillus subtilis membrane protein ComFA abolishes the uptake of DNA during transformation.
J Bacteriol: 1994, 176(15);4642-5
[PubMed:8045895]
[WorldCat.org]
[DOI]
(P p)
J A Londoño-Vallejo, D Dubnau
Membrane association and role in DNA uptake of the Bacillus subtilis PriA analogue ComF1.
Mol Microbiol: 1994, 13(2);197-205
[PubMed:7984101]
[WorldCat.org]
[DOI]
(P p)
J A Londoño-Vallejo, D Dubnau
comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases.
Mol Microbiol: 1993, 9(1);119-31
[PubMed:8412657]
[WorldCat.org]
[DOI]
(P p)