Difference between revisions of "ComEB"
| Line 60: | Line 60: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25580&redirect=T BSU25580] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/comEABC.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/comEABC.html] | ||
| Line 99: | Line 100: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25580&redirect=T BSU25580] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 15:16, 2 April 2014
- Description: late competence protein required for DNA binding and uptake
| Gene name | comEB |
| Synonyms | comD |
| Essential | no |
| Product | late competence protein |
| Function | genetic compentence |
| Gene expression levels in SubtiExpress: comEB | |
| Metabolic function and regulation of this protein in SubtiPathways: comEB | |
| MW, pI | 20 kDa, 5.368 |
| Gene length, protein length | 567 bp, 189 aa |
| Immediate neighbours | comEC, comEA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 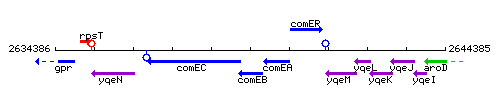
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed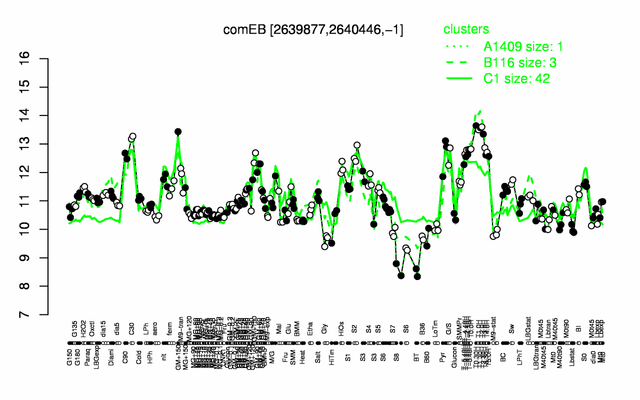
| |
Contents
Categories containing this gene/protein
genetic competence, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25580
Phenotypes of a mutant
Database entries
- BsubCyc: BSU25580
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: cytidine and deoxycytidylate deaminase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
- localizes to one or both cell poles PubMed
Database entries
- BsubCyc: BSU25580
- Structure:
- UniProt: P32393
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Miriam Kaufenstein, Martin van der Laan, Peter L Graumann
The three-layered DNA uptake machinery at the cell pole in competent Bacillus subtilis cells is a stable complex.
J Bacteriol: 2011, 193(7);1633-42
[PubMed:21278288]
[WorldCat.org]
[DOI]
(I p)
Mitsuo Ogura, Teruo Tanaka
The Bacillus subtilis late competence operon comE is transcriptionally regulated by yutB and under post-transcription initiation control by comN (yrzD).
J Bacteriol: 2009, 191(3);949-58
[PubMed:19028902]
[WorldCat.org]
[DOI]
(I p)
Hanne Jarmer, Randy Berka, Steen Knudsen, Hans H Saxild
Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions.
FEMS Microbiol Lett: 2002, 206(2);197-200
[PubMed:11814663]
[WorldCat.org]
[DOI]
(P p)
J Hahn, G Inamine, Y Kozlov, D Dubnau
Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA.
Mol Microbiol: 1993, 10(1);99-111
[PubMed:7968523]
[WorldCat.org]
[DOI]
(P p)