ClpX
- Description: ATP-dependent Clp protease ATP-binding subunit (class III heat-shock protein)
| Gene name | clpX |
| Synonyms | |
| Essential | no |
| Product | ATP-dependent Clp protease ATP-binding subunit |
| Function | protein degradation |
| Metabolic function and regulation of this protein in SubtiPathways: Phosphorelay, Stress | |
| MW, pI | 46 kDa, 4.645 |
| Gene length, protein length | 1260 bp, 420 aa |
| Immediate neighbours | lonB, tig |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 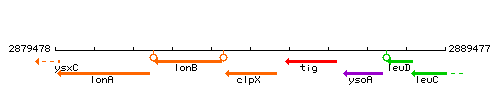
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU28220
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATPase/chaperone
- Protein family: clpX chaperone family (according to Swiss-Prot) ClpX (IP004487) InterPro, AAA+ -type ATPase (IPR013093) InterPro (PF07724) PFAM
Extended information on the protein
- Kinetic information:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
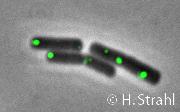
- Localization: cytoplasmic polar clusters, excluded from the nucleoid, induced clustering upon heat shock, colocalization with ClpP PubMed
Database entries
- Structure: homologue structure resolved 1UM8, structural model of B. subtilis ClpX available from hstrahl
- UniProt: P50866
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: clpX PubMed
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
Biological materials
- Mutant: clpX::kan, clpX::spec and clpX::cat available from the Hamoen] Lab
- Expression vector:
- lacZ fusion:
- GFP fusion: C-terminal GFP fusions (both single copy and 2th copy in amyE locus, also as CFP and YFP variants) available from the Hamoen] Lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Newcastle University, UK homepage
Your additional remarks
References
Reviews
David W Adams, Jeff Errington
Bacterial cell division: assembly, maintenance and disassembly of the Z ring.
Nat Rev Microbiol: 2009, 7(9);642-53
[PubMed:19680248]
[WorldCat.org]
[DOI]
(I p)
Janine Kirstein, Noël Molière, David A Dougan, Kürşad Turgay
Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases.
Nat Rev Microbiol: 2009, 7(8);589-99
[PubMed:19609260]
[WorldCat.org]
[DOI]
(I p)
Original Publications