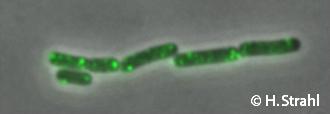
ClpE
- Description: ATP-dependent Clp protease-like (class III stress gene)
| Gene name | clpE |
| Synonyms | ykvH |
| Essential | no |
| Product | ATP-dependent Clp protease-like |
| Function | protein degradation |
| Gene expression levels in SubtiExpress: clpE | |
| Interactions involving this protein in SubtInteract: ClpE | |
| Metabolic function and regulation of this protein in SubtiPathways: clpE | |
| MW, pI | 77 kDa, 5.135 |
| Gene length, protein length | 2097 bp, 699 aa |
| Immediate neighbours | motA, ykvI |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 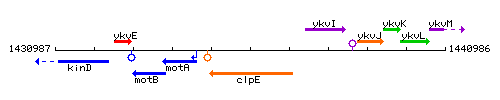
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed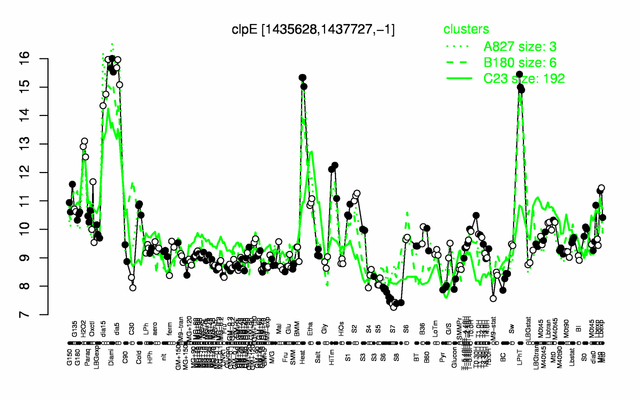
| |
Contents
Categories containing this gene/protein
proteolysis, heat shock proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU13700
Phenotypes of a mutant
Database entries
- BsubCyc: BSU13700
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATPase/chaperone
Extended information on the protein
- Kinetic information:
- Domains: AAA-ATPase PFAM
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU13700
- Structure:
- UniProt: O31673
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: clpE PubMed
- Additional information:
Biological materials
- Mutant: clpE::spec available from the Hamoen] Lab
- Expression vector:
- lacZ fusion:
- GFP fusion: C-terminal YFP and CFP fusions (single copy) available from the Hamoen] Lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Newcastle University, UK homepage
Your additional remarks
References
Reviews
Additional reviews: PubMed
Dorte Frees, Kirsi Savijoki, Pekka Varmanen, Hanne Ingmer
Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria.
Mol Microbiol: 2007, 63(5);1285-95
[PubMed:17302811]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Additional publications: PubMed
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
E Krüger, D Zühlke, E Witt, H Ludwig, M Hecker
Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor.
EMBO J: 2001, 20(4);852-63
[PubMed:11179229]
[WorldCat.org]
[DOI]
(P p)
I Derré, G Rapoport, K Devine, M Rose, T Msadek
ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis.
Mol Microbiol: 1999, 32(3);581-93
[PubMed:10320580]
[WorldCat.org]
[DOI]
(P p)
I Derré, G Rapoport, T Msadek
CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria.
Mol Microbiol: 1999, 31(1);117-31
[PubMed:9987115]
[WorldCat.org]
[DOI]
(P p)