Difference between revisions of "CitG"
| Line 122: | Line 122: | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 2427 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 2427 {{PubMed|24696501}} | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 9880 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 9880 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 7597 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 4282 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 4666 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
** GP718 (spec), available in [[Jörg Stülke]]'s lab | ** GP718 (spec), available in [[Jörg Stülke]]'s lab | ||
Revision as of 14:04, 17 April 2014
- Description: fumarase
| Gene name | citG |
| Synonyms | |
| Essential | no |
| Product | fumarate hydratase |
| Function | TCA cycle |
| Gene expression levels in SubtiExpress: citG | |
| Interactions involving this protein in SubtInteract: CitG | |
| Metabolic function and regulation of this protein in SubtiPathways: citG | |
| MW, pI | 50 kDa, 5.475 |
| Gene length, protein length | 1386 bp, 462 aa |
| Immediate neighbours | yuxN, yvzF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 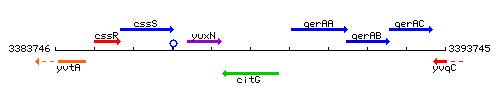
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed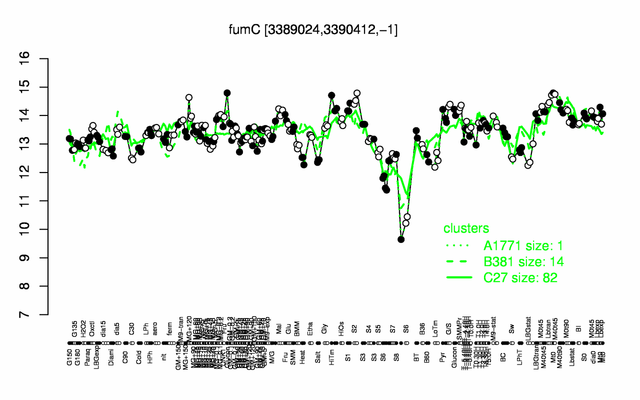
| |
Contents
Categories containing this gene/protein
carbon core metabolism, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33040
Phenotypes of a mutant
Database entries
- BsubCyc: BSU33040
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (S)-malate = fumarate + H2O (according to Swiss-Prot)
- Protein family: Fumarase subfamily (according to Swiss-Prot)
- Paralogous protein(s): AnsB
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU33040
- UniProt: P07343
- KEGG entry: [3]
- E.C. number: 4.2.1.2
Additional information
Expression and regulation
- Regulation: constitutive
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 2427 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 9880 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 7597 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 4282 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 4666 PubMed
Biological materials
- Mutant:
- GP718 (spec), available in Jörg Stülke's lab
- Expression vector:
- pGP1122 (N-terminal Strep-tag, for SPINE, purification from B. subtilis, in pGP380) (available in Jörg Stülke's lab)
- lacZ fusion:
- pGP387 (in pAC7), available in Jörg Stülke's lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
- FLAG-tag construct:
- GP1132 (spc, based on pGP1331), available in Jörg Stülke's lab
Labs working on this gene/protein
Your additional remarks
References
Guntur Fibriansah, Vinod Puthan Veetil, Gerrit J Poelarends, Andy-Mark W H Thunnissen
Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry: 2011, 50(27);6053-62
[PubMed:21661762]
[WorldCat.org]
[DOI]
(I p)
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
V A Price, I M Feavers, A Moir
Role of sigma H in expression of the fumarase gene (citG) in vegetative cells of Bacillus subtilis 168.
J Bacteriol: 1989, 171(11);5933-9
[PubMed:2509423]
[WorldCat.org]
[DOI]
(P p)
K M Tatti, H L Carter, A Moir, C P Moran
Sigma H-directed transcription of citG in Bacillus subtilis.
J Bacteriol: 1989, 171(11);5928-32
[PubMed:2509422]
[WorldCat.org]
[DOI]
(P p)
I M Feavers, V Price, A Moir
The regulation of the fumarase (citG) gene of Bacillus subtilis 168.
Mol Gen Genet: 1988, 211(3);465-71
[PubMed:3130545]
[WorldCat.org]
[DOI]
(P p)
J S Miles, J R Guest
Complete nucleotide sequence of the fumarase gene (citG) of Bacillus subtilis 168.
Nucleic Acids Res: 1985, 13(1);131-40
[PubMed:3923430]
[WorldCat.org]
[DOI]
(P p)