Difference between revisions of "CdsA"
| Line 58: | Line 58: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 86: | Line 83: | ||
* '''Interactions:''' | * '''Interactions:''' | ||
| − | * '''Localization:''' cell membrane {{PubMed|19820159}} | + | * '''Localization:''' cell membrane at the septum {{PubMed|15743965,19820159}} |
=== Database entries === | === Database entries === | ||
| Line 132: | Line 129: | ||
=References= | =References= | ||
| − | <pubmed>12884008,,19820159 </pubmed> | + | <pubmed>12884008,15743965,19820159 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:45, 30 March 2011
- Description: phosphatidate cytidylyltransferase
| Gene name | cdsA |
| Synonyms | |
| Essential | yes PubMed |
| Product | phosphatidate cytidylyltransferase |
| Function | biosynthesis of phospholipids |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis | |
| MW, pI | 30 kDa, 7.866 |
| Gene length, protein length | 807 bp, 269 aa |
| Immediate neighbours | uppS, ispC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 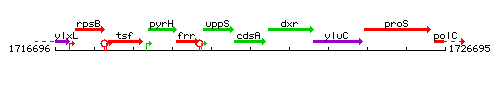
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
biosynthesis of lipids, essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16540
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: CTP + phosphatidate = diphosphate + CDP-diacylglycerol (according to Swiss-Prot)
- Protein family: CDS family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane at the septum PubMed
Database entries
- Structure:
- UniProt: O31752
- KEGG entry: [2]
- E.C. number: 2.7.7.41
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Jessica C Zweers, Thomas Wiegert, Jan Maarten van Dijl
Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis.
Appl Environ Microbiol: 2009, 75(23);7356-64
[PubMed:19820159]
[WorldCat.org]
[DOI]
(I p)
Ayako Nishibori, Jin Kusaka, Hiroshi Hara, Masato Umeda, Kouji Matsumoto
Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes.
J Bacteriol: 2005, 187(6);2163-74
[PubMed:15743965]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)