CdaR
- Description: effector protein controlling CdaA diadenylate cyclase activity
| Gene name | cdaR |
| Synonyms | ybbR |
| Essential | no |
| Product | effector protein controlling CdaA diadenylate cyclase activity |
| Function | regulation of c-di-AMP synthesis |
| Gene expression levels in SubtiExpress: cdaR | |
| Interactions involving this protein in SubtInteract: CdaR | |
| Metabolic function and regulation of this protein in SubtiPathways: cdaR | |
| MW, pI | 52 kDa, 4.719 |
| Gene length, protein length | 1449 bp, 483 aa |
| Immediate neighbours | cdaA, glmM |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 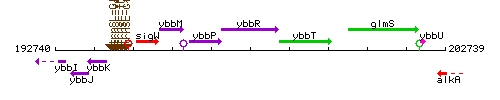
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
metabolism of signalling nucleotides
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01760
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O34659
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- GP999 (cdaR::cat), available in Jörg Stülke's lab
- GP985 (cdaA-cdaR::cat), available in Jörg Stülke's lab
- Expression vector:
- IPTG inducible expression of Strep-cdaR in E. coli: pGP2565 (in pGP172), available in Jörg Stülke's lab
- IPTG inducible expression of His-cdaR in E. coli: pGP2557 (in pET19b), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab. Respective plasmids: pGP1989 and pGP1992.
- Strep-tag construct:
- GP1330 (aphA3, based on pGP1460), available in Jörg Stülke's lab
- GP1380 lacA::cdaR-Strep aphA3 glmM-3xFLAG ermC (based on pGP1460 and pGP1087), available in Jörg Stülke's lab
- Antibody: available in Jörg Stülke's lab
Labs working on this gene/protein
Your additional remarks
References
Felix M P Mehne, Katrin Gunka, Hinnerk Eilers, Christina Herzberg, Volkhard Kaever, Jörg Stülke
Cyclic di-AMP homeostasis in bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth.
J Biol Chem: 2013, 288(3);2004-17
[PubMed:23192352]
[WorldCat.org]
[DOI]
(I p)
Yun Luo, John D Helmann
Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis.
Mol Microbiol: 2012, 83(3);623-39
[PubMed:22211522]
[WorldCat.org]
[DOI]
(I p)
Adam W Barb, John R Cort, Jayaraman Seetharaman, Scott Lew, Hsiau-Wei Lee, Thomas Acton, Rong Xiao, Michael A Kennedy, Liang Tong, Gaetano T Montelione, James H Prestegard
Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci: 2011, 20(2);396-405
[PubMed:21154411]
[WorldCat.org]
[DOI]
(I p)
Jun Yin, Paul D Straight, Shaun M McLoughlin, Zhe Zhou, Alison J Lin, David E Golan, Neil L Kelleher, Roberto Kolter, Christopher T Walsh
Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase.
Proc Natl Acad Sci U S A: 2005, 102(44);15815-20
[PubMed:16236721]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)