CcpC
| Gene name | ccpC |
| Synonyms | ykuM |
| Essential | no |
| Product | transcriptional repressor (LysR family) |
| Function | regulation of tricarboxylic acid branch of the TCA cycle |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 33 kDa, 5.932 |
| Gene length, protein length | 879 bp, 293 aa |
| Immediate neighbours | ykuL, ykuN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 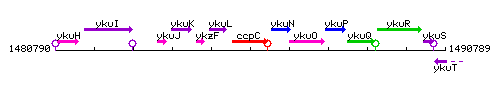
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed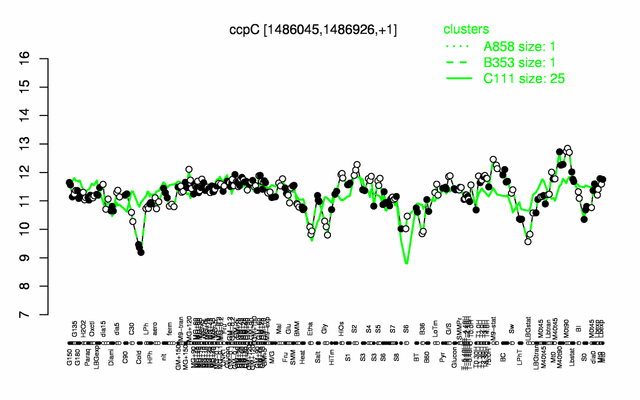
| |
Contents
Categories containing this gene/protein
carbon core metabolism, transcription factors and their control, regulators of core metabolism
This gene is a member of the following regulons
The CcpC regulon:
The gene
Basic information
- Locus tag: BSU14140
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: LysR family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O34827
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP206 (aphA3), available in Stülke lab
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Your additional remarks
References
Christina Herzberg, Lope Andrés Flórez Weidinger, Bastian Dörrbecker, Sebastian Hübner, Jörg Stülke, Fabian M Commichau
SPINE: a method for the rapid detection and analysis of protein-protein interactions in vivo.
Proteomics: 2007, 7(22);4032-5
[PubMed:17994626]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Irene Reif, Fabian M Commichau, Christian Detsch, Ingrid Wacker, Holger Ludwig, Jörg Stülke
Regulation of citB expression in Bacillus subtilis: integration of multiple metabolic signals in the citrate pool and by the general nitrogen regulatory system.
Arch Microbiol: 2006, 185(2);136-46
[PubMed:16395550]
[WorldCat.org]
[DOI]
(P p)
Sam-In Kim, Cécile Jourlin-Castelli, Stephen R Wellington, Abraham L Sonenshein
Mechanism of repression by Bacillus subtilis CcpC, a LysR family regulator.
J Mol Biol: 2003, 334(4);609-24
[PubMed:14636591]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Sam-In Kim, Manoja Ratnayake-Lecamwasam, Kiyoshi Tachikawa, Abraham L Sonenshein, Mark Strauch
Complex regulation of the Bacillus subtilis aconitase gene.
J Bacteriol: 2003, 185(5);1672-80
[PubMed:12591885]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Cécile Jourlin-Castelli, Sam-In Kim, Abraham L Sonenshein
Regulation of the bacillus subtilis ccpC gene by ccpA and ccpC.
Mol Microbiol: 2002, 43(2);399-410
[PubMed:11985717]
[WorldCat.org]
[DOI]
(P p)
C Jourlin-Castelli, N Mani, M M Nakano, A L Sonenshein
CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis.
J Mol Biol: 2000, 295(4);865-78
[PubMed:10656796]
[WorldCat.org]
[DOI]
(P p)