Difference between revisions of "CadA"
| Line 16: | Line 16: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU33490 cadA] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU33490 cadA] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=CadA CadA]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 75 kDa, 5.998 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 75 kDa, 5.998 | ||
Revision as of 11:53, 8 April 2014
- Description: cadmium transporting ATPase, resistance to cadmium
| Gene name | cadA |
| Synonyms | yvgW |
| Essential | no |
| Product | cadmium transporting ATPase |
| Function | cadmium export |
| Gene expression levels in SubtiExpress: cadA | |
| Metabolic function and regulation of this protein in SubtiPathways: CadA | |
| MW, pI | 75 kDa, 5.998 |
| Gene length, protein length | 2106 bp, 702 aa |
| Immediate neighbours | bdbD, copA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 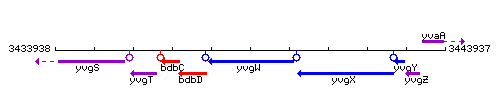
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed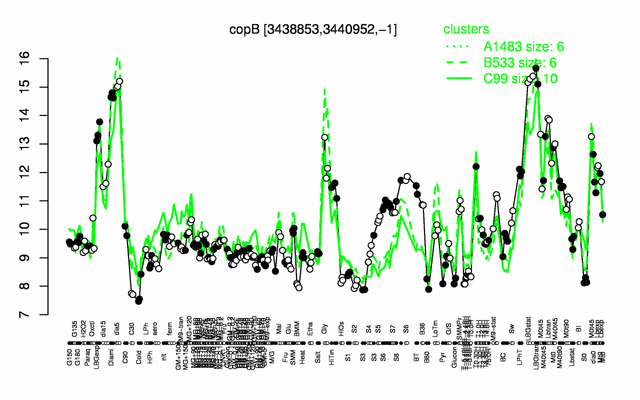
| |
Contents
Categories containing this gene/protein
transporters/ other, resistance against toxic metals, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33490
Phenotypes of a mutant
Database entries
- BsubCyc: BSU33490
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + H2O + Cd2+(In) = ADP + phosphate + Cd2+(Out) (according to Swiss-Prot)
- Protein family: Type IB subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- BsubCyc: BSU33490
- UniProt: O32219
- KEGG entry: [2]
- E.C. number: 3.6.3.3
Additional information
Expression and regulation
- Operon: cadA PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
Magnus Andersson, Daniel Mattle, Oleg Sitsel, Tetyana Klymchuk, Anna Marie Nielsen, Lisbeth Birk Møller, Stephen H White, Poul Nissen, Pontus Gourdon
Copper-transporting P-type ATPases use a unique ion-release pathway.
Nat Struct Mol Biol: 2014, 21(1);43-8
[PubMed:24317491]
[WorldCat.org]
[DOI]
(I p)
Charles M Moore, Ahmed Gaballa, Monica Hui, Rick W Ye, John D Helmann
Genetic and physiological responses of Bacillus subtilis to metal ion stress.
Mol Microbiol: 2005, 57(1);27-40
[PubMed:15948947]
[WorldCat.org]
[DOI]
(P p)
Ahmed Gaballa, John D Helmann
Bacillus subtilis CPx-type ATPases: characterization of Cd, Zn, Co and Cu efflux systems.
Biometals: 2003, 16(4);497-505
[PubMed:12779235]
[WorldCat.org]
[DOI]
(P p)
I M Solovieva, K D Entian
Investigation of the yvgW Bacillus subtilis chromosomal gene involved in Cd(2+) ion resistance.
FEMS Microbiol Lett: 2002, 208(1);105-9
[PubMed:11934502]
[WorldCat.org]
[DOI]
(P p)