Difference between revisions of "Buk"
| Line 60: | Line 60: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU24070&redirect=T BSU24070] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ptb-bcd-buk-lpdV-bkdAABB.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ptb-bcd-buk-lpdV-bkdAABB.html] | ||
| Line 97: | Line 98: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU24070&redirect=T BSU24070] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?pdbId=1X9J 1X9J] (from ''Thermotoga maritima'', 52% identity, 69% similarity) | * '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?pdbId=1X9J 1X9J] (from ''Thermotoga maritima'', 52% identity, 69% similarity) | ||
Revision as of 14:10, 2 April 2014
- Description: butyrate kinase
| Gene name | buk |
| Synonyms | yqiU, bkd |
| Essential | no |
| Product | butyrate kinase |
| Function | utilization of branched-chain keto acids |
| Gene expression levels in SubtiExpress: buk | |
| Metabolic function and regulation of this protein in SubtiPathways: buk | |
| MW, pI | 39 kDa, 5.466 |
| Gene length, protein length | 1089 bp, 363 aa |
| Immediate neighbours | lpdV, bcd |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 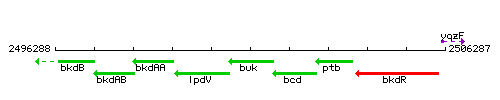
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
BkdR regulon, CodY regulon, SigL regulon
The gene
Basic information
- Locus tag: BSU24070
Phenotypes of a mutant
Database entries
- BsubCyc: BSU24070
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + butanoate = ADP + butanoyl phosphate (according to Swiss-Prot)
- Protein family: acetokinase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU24070
- Structure: 1X9J (from Thermotoga maritima, 52% identity, 69% similarity)
- UniProt: P54532
- KEGG entry: [3]
- E.C. number: 2.7.2.7
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
M Nickel, G Homuth, C Böhnisch, U Mäder, T Schweder
Cold induction of the Bacillus subtilis bkd operon is mediated by increased mRNA stability.
Mol Genet Genomics: 2004, 272(1);98-107
[PubMed:15241682]
[WorldCat.org]
[DOI]
(P p)
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)
Tanja Kaan, Georg Homuth, Ulrike Mäder, Julia Bandow, Thomas Schweder
Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response.
Microbiology (Reading): 2002, 148(Pt 11);3441-3455
[PubMed:12427936]
[WorldCat.org]
[DOI]
(P p)
M Debarbouille, R Gardan, M Arnaud, G Rapoport
Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis.
J Bacteriol: 1999, 181(7);2059-66
[PubMed:10094682]
[WorldCat.org]
[DOI]
(P p)