Difference between revisions of "BsrA"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || antagonist of [[SigA]] | |style="background:#ABCDEF;" align="center"|'''Function''' || antagonist of [[SigA]] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this RNA in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this RNA in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=BsrA BsrA] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Gene length''' || 190 bp | |style="background:#ABCDEF;" align="center"| '''Gene length''' || 190 bp | ||
Revision as of 14:12, 18 November 2013
- Description: 6S RNA, important for timing of sporulation
| Gene name | bsrA |
| Synonyms | |
| Essential | no |
| Product | 6S RNA |
| Function | antagonist of SigA |
| Interactions involving this RNA in SubtInteract: BsrA | |
| Gene length | 190 bp |
| Immediate neighbours | yrvM, aspS |
| Gene sequence (+200bp) | |
Genetic context 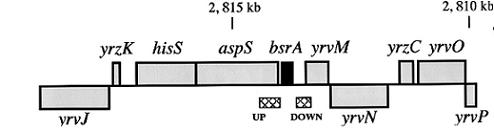
| |
Contents
Categories containing this gene/protein
sigma factors and their control, ncRNA
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU_misc_RNA_41
Phenotypes of a mutant
- reduced growth rate PubMed
- delayed outgrowth from stationary phase PubMed
- altered timing of onset of sporulation (earlier sporulation) PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: no entry
- Structure:
Additional information
- Similar RNA: bsrB
Expression and regulation
- Operon: bsrA PubMed
- Regulation: expressed in vegetative cells PubMed
- Regulatory mechanism:
- Additional information: transcribed as 201 nt RNA, processed to mature 190 bp 6S RNA PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
Labs working on this gene/RNA
Kouji Nakamura, Tsukuba, University, Japan
Roland Hartmann, Philipps-Universität Marburg, Marburg, Germany, Homepage
Your additional remarks
References
Ignacio J Cabrera-Ostertag, Amy T Cavanagh, Karen M Wassarman
Initiating nucleotide identity determines efficiency of RNA synthesis from 6S RNA templates in Bacillus subtilis but not Escherichia coli.
Nucleic Acids Res: 2013, 41(15);7501-11
[PubMed:23761441]
[WorldCat.org]
[DOI]
(I p)
Amy T Cavanagh, Karen M Wassarman
6S-1 RNA function leads to a delay in sporulation in Bacillus subtilis.
J Bacteriol: 2013, 195(9);2079-86
[PubMed:23457253]
[WorldCat.org]
[DOI]
(I p)
Benedikt M Beckmann, Philipp G Hoch, Manja Marz, Dagmar K Willkomm, Margarita Salas, Roland K Hartmann
A pRNA-induced structural rearrangement triggers 6S-1 RNA release from RNA polymerase in Bacillus subtilis.
EMBO J: 2012, 31(7);1727-38
[PubMed:22333917]
[WorldCat.org]
[DOI]
(I p)
Amy T Cavanagh, Jamie M Sperger, Karen M Wassarman
Regulation of 6S RNA by pRNA synthesis is required for efficient recovery from stationary phase in E. coli and B. subtilis.
Nucleic Acids Res: 2012, 40(5);2234-46
[PubMed:22102588]
[WorldCat.org]
[DOI]
(I p)
Benedikt M Beckmann, Olga Y Burenina, Philipp G Hoch, Elena A Kubareva, Cynthia M Sharma, Roland K Hartmann
In vivo and in vitro analysis of 6S RNA-templated short transcripts in Bacillus subtilis.
RNA Biol: 2011, 8(5);839-49
[PubMed:21881410]
[WorldCat.org]
[DOI]
(I p)
Jeffrey E Barrick, Narasimhan Sudarsan, Zasha Weinberg, Walter L Ruzzo, Ronald R Breaker
6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter.
RNA: 2005, 11(5);774-84
[PubMed:15811922]
[WorldCat.org]
[DOI]
(P p)
Satoru Suzuma, Sayaka Asari, Keigo Bunai, Keiko Yoshino, Yoshinari Ando, Hiroshi Kakeshita, Masaya Fujita, Kouji Nakamura, Kunio Yamane
Identification and characterization of novel small RNAs in the aspS-yrvM intergenic region of the Bacillus subtilis genome.
Microbiology (Reading): 2002, 148(Pt 8);2591-2598
[PubMed:12177353]
[WorldCat.org]
[DOI]
(P p)