Difference between revisions of "BofA"
| Line 31: | Line 31: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=bofA_29772_30035_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:bofA_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=bofA_29772_30035_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:bofA_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU00230]] |
|- | |- | ||
|} | |} | ||
Revision as of 12:17, 16 May 2013
- Description: inhibition of the pro-sigma-K processing machinery
| Gene name | bofA |
| Synonyms | |
| Essential | no |
| Product | inhibitor of pro-SigK processing |
| Function | control of SigK activation |
| Gene expression levels in SubtiExpress: bofA | |
| Interactions involving this protein in SubtInteract: BofA | |
| MW, pI | 8 kDa, 9.32 |
| Gene length, protein length | 261 bp, 87 aa |
| Immediate neighbours | yaaL, rrnA-16S |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 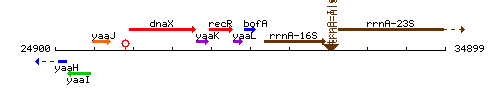
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed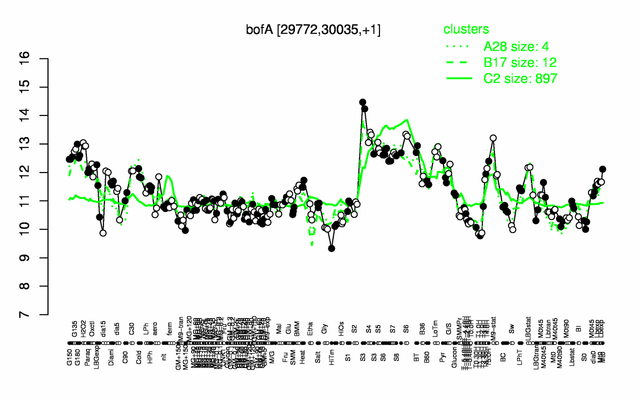
| |
Contents
Categories containing this gene/protein
sigma factors and their control, sporulation proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00230
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: forespore outer membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P24282
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: bofA PubMed
- Regulation: expressed during sporulation PubMed
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Simon Cutting, London, UK homepage
Your additional remarks
References
Nathalie Campo, David Z Rudner
A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis.
Mol Cell: 2006, 23(1);25-35
[PubMed:16818230]
[WorldCat.org]
[DOI]
(P p)
Thierry Doan, Kathleen A Marquis, David Z Rudner
Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum.
Mol Microbiol: 2005, 55(6);1767-81
[PubMed:15752199]
[WorldCat.org]
[DOI]
(P p)
Tran C Dong, Simon M Cutting
The PDZ domain of the SpoIVB transmembrane signaling protein enables cis-trans interactions involving multiple partners leading to the activation of the pro-sigmaK processing complex in Bacillus subtilis.
J Biol Chem: 2004, 279(42);43468-78
[PubMed:15292188]
[WorldCat.org]
[DOI]
(P p)
Tran C Dong, Simon M Cutting
SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-sigmaK in Bacillus subtilis.
Mol Microbiol: 2003, 49(5);1425-34
[PubMed:12940997]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
David Z Rudner, Richard Losick
A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization.
Genes Dev: 2002, 16(8);1007-18
[PubMed:11959848]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, R Losick
Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1998, 95(6);3162-7
[PubMed:9501233]
[WorldCat.org]
[DOI]
(P p)
E Ricca, S Cutting, R Losick
Characterization of bofA, a gene involved in intercompartmental regulation of pro-sigma K processing during sporulation in Bacillus subtilis.
J Bacteriol: 1992, 174(10);3177-84
[PubMed:1577688]
[WorldCat.org]
[DOI]
(P p)
K Ireton, A D Grossman
Interactions among mutations that cause altered timing of gene expression during sporulation in Bacillus subtilis.
J Bacteriol: 1992, 174(10);3185-95
[PubMed:1315731]
[WorldCat.org]
[DOI]
(P p)
S Cutting, V Oke, A Driks, R Losick, S Lu, L Kroos
A forespore checkpoint for mother cell gene expression during development in B. subtilis.
Cell: 1990, 62(2);239-50
[PubMed:2115401]
[WorldCat.org]
[DOI]
(P p)