Difference between revisions of "BglS"
| Line 20: | Line 20: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[citH]]'', ''[[licT]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[citH]]'', ''[[licT]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''[http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB15943]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
| − | |||
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:bglS_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:bglS_context.gif]] | ||
Revision as of 00:30, 17 April 2009
- Description: endo-beta-1,3-1,4 glucanase
| Gene name | bglS |
| Synonyms | bgl, licS |
| Essential | no |
| Product | endo-beta-1,3-1,4 glucanase |
| Function | lichenan degradation |
| MW, pI | 27 kDa, 6.482 |
| Gene length, protein length | 726 bp, 242 aa |
| Immediate neighbours | citH, licT |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 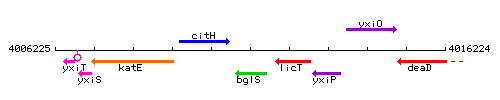
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: extracellular (signal peptide) PubMed, extracellular
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [3]
- E.C. number: 3.2.1.73
Additional information
Expression and regulation
- Regulation: repressed by glucose (3.1-fold) (CcpA) PubMed, expressed in the stationary phase (temporal activation), carbon catabolite repression (CcpA) PubMed, weak induction by ß-glucosides (LicT) PubMed
- Regulatory mechanism: CcpA: transcription repression, CcpA: transcription repression PubMed, LicT: binding to an RNA switch results in transcriptional antitermination PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Voigt et al. (2009) Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis. J Mol Microbiol Biotechnol. 16: 53-68 PubMed
- Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng. 5: 133-149 PubMed
- Murphy ND, McConnell DJ, Cantwell BA. 1984. The DNA sequence of the gene and genetic control sites for the excreted Bacillus subtilis enzyme β-glucanase. Nucl. Acids Res. 12:5355-5367. PubMed
- Stülke, J., Hanschke, R. & Hecker, M. (1993) Temporal activation of ß-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139: 2041-2045. PubMed
- Krüger, S., Stülke, J. & Hecker, M. (1993) Carbon catabolite repression of ß-glucanase synthesis in Bacillus subtilis. J. Gen. Microbiol. 139: 2047-2054. PubMed
- Schnetz, K., Stülke, J., Gertz, S., Krüger, S., Krieg, M., Hecker, M. & Rak, B. (1996) LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J. Bacteriol. 178: 1971-1979. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed