Difference between revisions of "BglC"
| Line 125: | Line 125: | ||
** 1A751 (no resistance), {{PubMed|7704256}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A751&Search=1A751 BGSC] | ** 1A751 (no resistance), {{PubMed|7704256}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A751&Search=1A751 BGSC] | ||
** 1A752 (no resistance), {{PubMed|7704256}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A752&Search=1A752 BGSC] | ** 1A752 (no resistance), {{PubMed|7704256}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A752&Search=1A752 BGSC] | ||
| + | ** 1A976 (no resistance), {{PubMed|21255377}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A976&Search=1A976 BGSC] | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
Revision as of 10:42, 20 September 2012
- Description: endo-1,4-beta-glucanase
| Gene name | bglC |
| Synonyms | eglS |
| Essential | no |
| Product | endo-1,4-beta-glucanase) |
| Function | beta-1,4-glucan degradation |
| Gene expression levels in SubtiExpress: bglC | |
| MW, pI | 55 kDa, 8.619 |
| Gene length, protein length | 1497 bp, 499 aa |
| Immediate neighbours | alsT, ynfE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 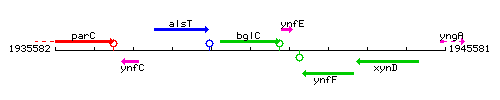
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed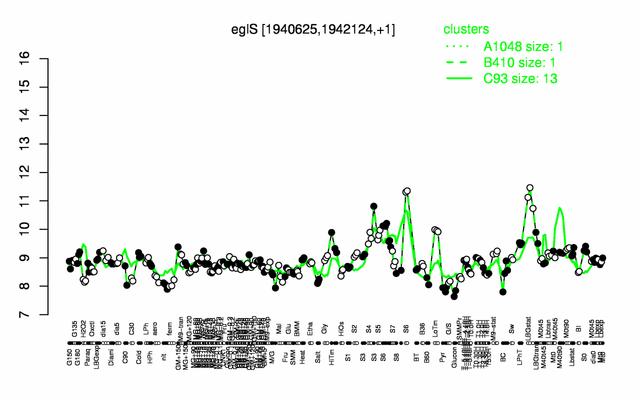
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU18130
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Endohydrolysis of (1->4)-beta-D-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 5 (cellulase A) family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: extracellular (signal peptide) PubMed
Database entries
- UniProt: P10475
- KEGG entry: [3]
- E.C. number: 3.2.1.4
Additional information
Expression and regulation
- Operon: bglC (according to DBTBS)
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Camila R Santos, Joice H Paiva, Maurício L Sforça, Jorge L Neves, Rodrigo Z Navarro, Júnio Cota, Patrícia K Akao, Zaira B Hoffmam, Andréia N Meza, Juliana H Smetana, Maria L Nogueira, Igor Polikarpov, José Xavier-Neto, Fábio M Squina, Richard J Ward, Roberto Ruller, Ana C Zeri, Mário T Murakami
Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem J: 2012, 441(1);95-104
[PubMed:21880019]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
L M Robson, G H Chambliss
Endo-beta-1,4-glucanase gene of Bacillus subtilis DLG.
J Bacteriol: 1987, 169(5);2017-25
[PubMed:3106328]
[WorldCat.org]
[DOI]
(P p)
R M MacKay, A Lo, G Willick, M Zuker, S Baird, M Dove, F Moranelli, V Seligy
Structure of a Bacillus subtilis endo-beta-1,4-glucanase gene.
Nucleic Acids Res: 1986, 14(22);9159-70
[PubMed:3024130]
[WorldCat.org]
[DOI]
(P p)