Difference between revisions of "BglA"
| Line 119: | Line 119: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 230 {{PubMed|21395229}} | ||
| + | |||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 362 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
Revision as of 13:33, 17 April 2014
- Description: 6-phospho-beta-glucosidase
| Gene name | bglA |
| Synonyms | |
| Essential | no |
| Product | 6-phospho-beta-glucosidase) |
| Function | beta-glucoside utilization |
| Gene expression levels in SubtiExpress: bglA | |
| MW, pI | 54 kDa, 4.648 |
| Gene length, protein length | 1437 bp, 479 aa |
| Immediate neighbours | ahpF, yyzE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 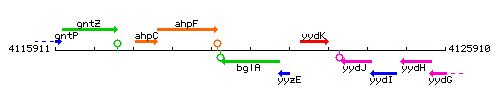
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed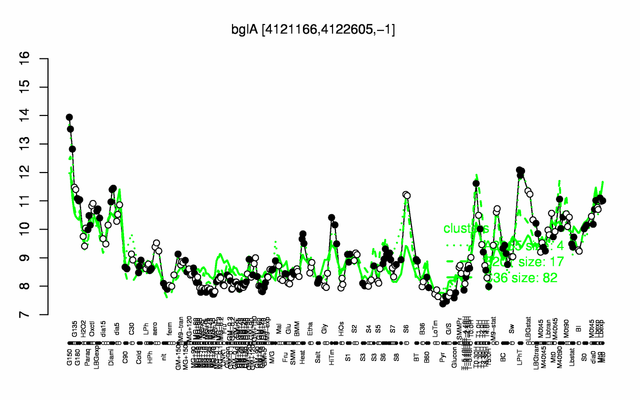
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40110
Phenotypes of a mutant
Database entries
- BsubCyc: BSU40110
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 6-phospho-beta-D-glucosyl-(1,4)-D-glucose + H2O = D-glucose + D-glucose 6-phosphate (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 1 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU40110
- Structure:
- UniProt: P42973
- KEGG entry: [3]
- E.C. number: 3.2.1.86
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 230 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 362 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
B Setlow, R-M Cabrera-Martinez, P Setlow
Mechanism of the hydrolysis of 4-methylumbelliferyl-beta-D-glucoside by germinating and outgrowing spores of Bacillus species.
J Appl Microbiol: 2004, 96(6);1245-55
[PubMed:15139916]
[WorldCat.org]
[DOI]
(P p)
Barbara Setlow, Arturo Cabrera-Hernandez, Rosa Martha Cabrera-Martinez, Peter Setlow
Identification of aryl-phospho-beta-D-glucosidases in Bacillus subtilis.
Arch Microbiol: 2004, 181(1);60-7
[PubMed:14652714]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)
J Zhang, A Aronson
A Bacillus subtilis bglA gene encoding phospho-beta-glucosidase is inducible and closely linked to a NADH dehydrogenase-encoding gene.
Gene: 1994, 140(1);85-90
[PubMed:8125345]
[WorldCat.org]
[DOI]
(P p)