Difference between revisions of "BdhA"
| Line 34: | Line 34: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 111: | Line 107: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=bdhA_677911_678951_-1 bdhA] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=bdhA_677911_678951_-1 bdhA] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 19:38, 18 June 2013
- Description: acetoine/ butanediol dehydrogenase
| Gene name | bdhA |
| Synonyms | ydjL |
| Essential | no |
| Product | acetoine/ butanediol dehydrogenase |
| Function | overflow metabolism, fermentation |
| Gene expression levels in SubtiExpress: bdhA | |
| MW, pI | 37 kDa, 4.805 |
| Gene length, protein length | 1038 bp, 346 aa |
| Immediate neighbours | iolT, ydjM |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 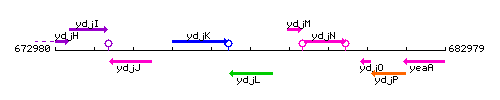
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed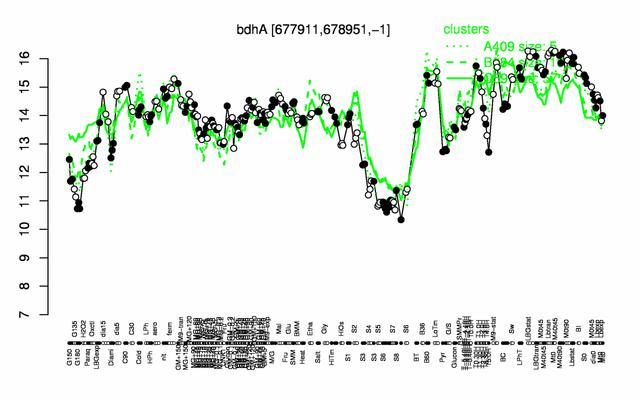
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06240
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: formation of butanediol from acetoin PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-13 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O34788
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: bdhA (according to DBTBS)
- Regulation:
- Additional information:
- The mRNA has a long (268 nt) untranslated leader region PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Rafael R de Oliveira, Wayne L Nicholson
The LysR-type transcriptional regulator (LTTR) AlsR indirectly regulates expression of the Bacillus subtilis bdhA gene encoding 2,3-butanediol dehydrogenase.
Appl Microbiol Biotechnol: 2013, 97(16);7307-16
[PubMed:23576037]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Ranjita Biswas, Masaru Yamaoka, Hideki Nakayama, Takashi Kondo, Ken-ichi Yoshida, Virendra S Bisaria, Akihiko Kondo
Enhanced production of 2,3-butanediol by engineered Bacillus subtilis.
Appl Microbiol Biotechnol: 2012, 94(3);651-8
[PubMed:22361854]
[WorldCat.org]
[DOI]
(I p)
Claudia Frädrich, Anika March, Kerstin Fiege, Anja Hartmann, Dieter Jahn, Elisabeth Härtig
The transcription factor AlsR binds and regulates the promoter of the alsSD operon responsible for acetoin formation in Bacillus subtilis.
J Bacteriol: 2012, 194(5);1100-12
[PubMed:22178965]
[WorldCat.org]
[DOI]
(I p)
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Wayne L Nicholson
The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase.
Appl Environ Microbiol: 2008, 74(22);6832-8
[PubMed:18820069]
[WorldCat.org]
[DOI]
(I p)