Difference between revisions of "BacB"
| Line 59: | Line 59: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU37730&redirect=T BSU37730] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 93: | Line 94: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU37730&redirect=T BSU37730] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3H7J 3H7J] {{PubMed|19776011}} | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3H7J 3H7J] {{PubMed|19776011}} | ||
Revision as of 15:06, 2 April 2014
- Description: oxidase that catalyzes the synthesis of 2-oxo-3-(4-oxocyclohexa-2,5-dienyl)propanoic acid, a precursor to L-anticapsin
| Gene name | bacB |
| Synonyms | ywfC, ipa-81d |
| Essential | no |
| Product | oxidase |
| Function | biosynthesis of the antibiotic bacilysin |
| Gene expression levels in SubtiExpress: bacB | |
| MW, pI | 26 kDa, 5.193 |
| Gene length, protein length | 705 bp, 235 aa |
| Immediate neighbours | bacC, bacA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 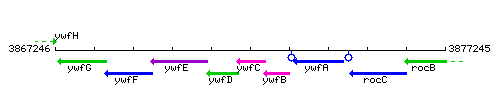
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed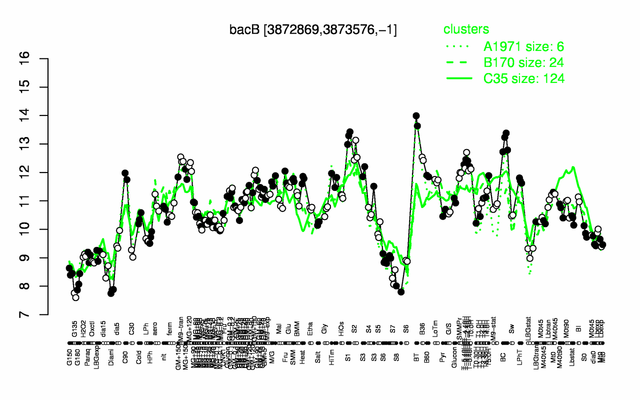
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds
This gene is a member of the following regulons
AbrB regulon, CodY regulon, ScoC regulon
The gene
Basic information
- Locus tag: BSU37730
Phenotypes of a mutant
Database entries
- BsubCyc: BSU37730
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: synthesis of 2-oxo-3-(4-oxocyclohexa-2,5-dienyl)propanoic acid, a precursor to L-anticapsin PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU37730
- UniProt: P39639
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Takashi Inaoka, Kozo Ochi
Scandium stimulates the production of amylase and bacilysin in Bacillus subtilis.
Appl Environ Microbiol: 2011, 77(22);8181-3
[PubMed:21948839]
[WorldCat.org]
[DOI]
(I p)
M Rajavel, B Gopal
Analysis of multiple crystal forms of Bacillus subtilis BacB suggests a role for a metal ion as a nucleant for crystallization.
Acta Crystallogr D Biol Crystallogr: 2010, 66(Pt 5);635-9
[PubMed:20445239]
[WorldCat.org]
[DOI]
(I p)
Sarah A Mahlstedt, Christopher T Walsh
Investigation of anticapsin biosynthesis reveals a four-enzyme pathway to tetrahydrotyrosine in Bacillus subtilis.
Biochemistry: 2010, 49(5);912-23
[PubMed:20052993]
[WorldCat.org]
[DOI]
(I p)
Takashi Inaoka, Guojun Wang, Kozo Ochi
ScoC regulates bacilysin production at the transcription level in Bacillus subtilis.
J Bacteriol: 2009, 191(23);7367-71
[PubMed:19801406]
[WorldCat.org]
[DOI]
(I p)
Malligarjunan Rajavel, Ashima Mitra, Balasubramanian Gopal
Role of Bacillus subtilis BacB in the synthesis of bacilysin.
J Biol Chem: 2009, 284(46);31882-92
[PubMed:19776011]
[WorldCat.org]
[DOI]
(I p)
Gerhard Steinborn, Mohammad-Reza Hajirezaei, Jürgen Hofemeister
bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains.
Arch Microbiol: 2005, 183(2);71-9
[PubMed:15609023]
[WorldCat.org]
[DOI]
(P p)
Takashi Inaoka, Kosaku Takahashi, Mayumi Ohnishi-Kameyama, Mitsuru Yoshida, Kozo Ochi
Guanine nucleotides guanosine 5'-diphosphate 3'-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis.
J Biol Chem: 2003, 278(4);2169-76
[PubMed:12372825]
[WorldCat.org]
[DOI]
(P p)