Difference between revisions of "ArtP"
(→References) |
|||
| Line 80: | Line 80: | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2Q2C 2Q2C] (Geobacillus stearothermophilus) | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2Q2C 2Q2C] (Geobacillus stearothermophilus) | ||
| − | * ''' | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/P54535 P54535] |
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU23980] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU23980] | ||
Revision as of 13:29, 20 July 2009
- Description: high affinity arginine ABC transporter (ATP-binding protein)
| Gene name | artP |
| Synonyms | yqiX |
| Essential | no |
| Product | high affinity arginine ABC transporter (ATP-binding protein) |
| Function | arginine uptake |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 28 kDa, 5.086 |
| Gene length, protein length | 765 bp, 255 aa |
| Immediate neighbours | artQ, yqiW |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 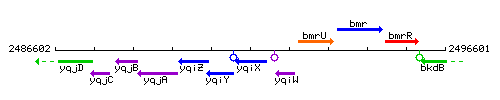
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU23980
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: bacterial solute-binding protein 3 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot), extracellular (signal peptide) PubMed, membrane PubMed
Database entries
- Structure: 2Q2C (Geobacillus stearothermophilus)
- UniProt: P54535
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation: repressed by casamino acids PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
A Sekowska, S Robin, J J Daudin, A Hénaut, A Danchin
Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis.
Genome Biol: 2001, 2(6);RESEARCH0019
[PubMed:11423008]
[WorldCat.org]
[DOI]
(I p)
Y Quentin, G Fichant, F Denizot
Inventory, assembly and analysis of Bacillus subtilis ABC transport systems.
J Mol Biol: 1999, 287(3);467-84
[PubMed:10092453]
[WorldCat.org]
[DOI]
(P p)