Difference between revisions of "AroK"
(→References) |
|||
| Line 16: | Line 16: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU03150 aroK] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU03150 aroK] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=aroK aroK]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 5.969 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 5.969 | ||
Revision as of 11:07, 7 January 2014
- Description: shikimate kinase
| Gene name | aroK |
| Synonyms | aroI |
| Essential | no |
| Product | shikimate kinase |
| Function | biosynthesis of aromatic amino acids |
| Gene expression levels in SubtiExpress: aroK | |
| Metabolic function and regulation of this protein in SubtiPathways: aroK | |
| MW, pI | 21 kDa, 5.969 |
| Gene length, protein length | 558 bp, 186 aa |
| Immediate neighbours | tmrB, ycgJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 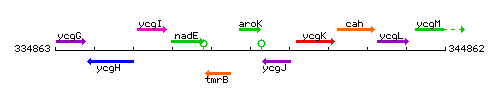
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed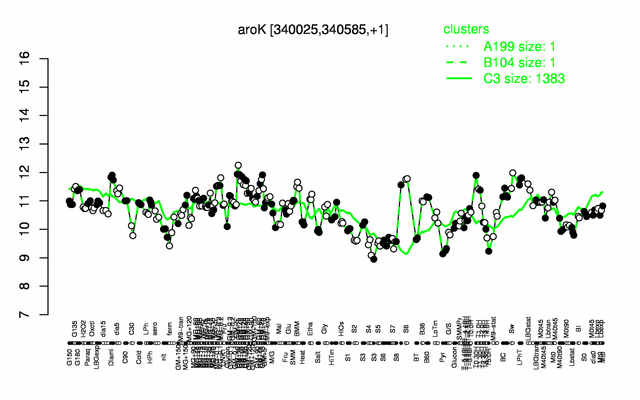
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU03150
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + shikimate = ADP + shikimate 3-phosphate (according to Swiss-Prot)
- Protein family: shikimate kinase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P37944
- KEGG entry: [3]
- E.C. number: 2.7.1.71
Additional information
Expression and regulation
- Operon: aroK (according to DBTBS)
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Cuauhtemoc Licona-Cassani, Alvaro R Lara, Natividad Cabrera-Valladares, Adelfo Escalante, Georgina Hernández-Chávez, Alfredo Martinez, Francisco Bolívar, Guillermo Gosset
Inactivation of pyruvate kinase or the phosphoenolpyruvate: sugar phosphotransferase system increases shikimic and dehydroshikimic acid yields from glucose in Bacillus subtilis.
J Mol Microbiol Biotechnol: 2014, 24(1);37-45
[PubMed:24158146]
[WorldCat.org]
[DOI]
(I p)
L Huang, A L Montoya, E W Nester
Purification and characterization of shikimate kinase enzyme activity in Bacillus subtilis.
J Biol Chem: 1975, 250(19);7675-81
[PubMed:170268]
[WorldCat.org]
(P p)
J A Hoch, E W Nester
Gene-enzyme relationships of aromatic acid biosynthesis in Bacillus subtilis.
J Bacteriol: 1973, 116(1);59-66
[PubMed:4200844]
[WorldCat.org]
[DOI]
(P p)
E W Nester, R A Jensen, D S Nasser
Regulation of enzyme synthesis in the aromatic amino acid pathway of Bacillus subtilus.
J Bacteriol: 1969, 97(1);83-90
[PubMed:4974400]
[WorldCat.org]
[DOI]
(P p)